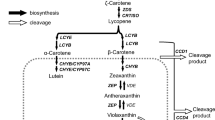
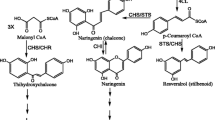
Abstract
Recent studies on chalcone synthase (CHS) and the related stilbene synthase (STS) suggest that the structure of chs-like genes in plants has evolved into different forms, whose members have both different regulation and capacity to code for different but related enzymatic activities. We have studied the diversity of chs-like genes by analysing the structure, expression patterns and catalytic properties of the corresponding enzymes of three genes that are active during corolla development in Gerbera hybrida. The expression patterns demonstrate that chs-like genes are representatives of three distinct genetic programmes that are active during organ differentiation in gerbera. Gchs1 and gchs3 code for typical CHS enzymes, and their gene expression pattern temporally correlates with flavonol (gchs1, gchs3) and anthocyanin (gchs1) synthesis during corolla development. Gchs2 is different. The expression pattern does not correlate with the pigmentation pattern, the amino acid sequence deviates considerably from the consensus of typical CHSs, and the catalytic properties are different. The data indicate that it represents a new member in the large superfamily of chs and chs-related genes.
Similar content being viewed by others
References
Asen S: High pressure liquid chromatographic analysis of flavonoid chemical markers in petals from Gerbera flowers as an adjunct for cultivar and germplasm identification. Phytochemistry 23: 2523–2526 (1984).
Batschauer A, Ehmann B, Schäfer E: Cloning and characterization of a chalcone synthase gene from mustard and its light-dependent expression. Plant Mol Biol 16: 175–185 (1991).
Baumert A, Maier W, Groeger D, Deutzmann R: Purification and properties of acridone synthase from cell suspension cultures of Ruta graveolens. Z Naturforsch 49c: 26–32 (1994).
Bohlmann F, Grenz M: Über die Inhaltsstoffe von Gerbera piloselloides Cass. Chem Ber 108: 26–30 (1975).
Borejsza-Wysocki W, Hrazdina G: Biosynthesis of p-hydroxyphenylbutan-2-one in raspberry fruits and tissue cultures. Phytochemistry 35: 623–628 (1994).
Bowers WS, Ohta T, Cleere JS, Marsella PA: Discovery of insect anti-juvenile hormones in plants. Science 193: 542–547 (1976).
Coe EH, McCormick S, Modena SA: White pollen in maize. J Hered 72: 318–320 (1981).
Coen E, Meyerowitz EMM: The war of whorls: genetic interactions controlling flower development. Nature 353: 31–37 (1991).
Cox KH, Goldberg RB: Analysis of plant gene expression. In: Shaw CH (ed) Plant Molecular Biology: A Practical Approach, pp. 1–34. IRL Press, Oxford (1988).
Dangl JL, Hahlbrock K, Schell J: Regulation and structure of chalcone synthase genes. In: Vasil IK, Schell J (eds) Cell Culture and Somatic Cell Genetics of Plants 4, pp. 155–173. Academic Press, New York (1989).
Davies KM, Bradley JM, Schwinn KE, Markham KR, Podivinsky E: Flavonoid biosynthesis in flower petals of five lines of lisianthus (Eustoma grandiflorum Grise.). Plant Sci 95: 67–77 (1993).
Dellaporta SL, Wood J, Hicks JB: A plant DNA mini-preparation: Version II. Plant Mol Biol Rep 1: 19–21 (1983).
Ehmann B, Schäfer E: Nucleotide sequences encoding two different chalcone synthases expressed in cotyledons of SAN 9789 treated mustard (Sinapis alba L.). Plant Mol Biol 11: 869–870 (1988).
Elomaa P, Honkanen J, Puska R, Seppänen P, Helariutta Y, Mehto M, Kotilainen M, Nevalainen L, Teeri TH: Agrobacterium-mediated transfer of antisense chalcone synthase cDNA to Gerbera hybrida inhibits flower pigmentation. Bio/technology 11: 508–511 (1993).
Feinbaum RL, Ausubel FM: Transcriptional regulation of the Arabidopsis thaliana chalcone synthase gene. Mol Cell Biol 8: 1985–1992 (1988).
Fliegmann J, Schröder G, Schanz S, Britsch L, Schröder J: Molecular analysis of chalcone and dihydropinosylvin synthase from Scots pine (Pinus sylvestris), and differential regulation of these and related activities in stressed plants. Plant Mol Biol 18: 489–503 (1992).
Franken P, Niesbach-Klösgen U, Weydemann U, Marechal-Drouard L, Saedler H, Wienand U: The duplicated c chalcone synthase genes C2 and Whp (white pollen) of Zea mays are independently regulated; evidence for translational control of Whp expression by the anthocyanin intensifying gene in. EMBO J 10: 2605–2612 (1991).
Gerats AGM, De Vlaming P, Doodeman M, Al B, Schram AW: Genetic control of the conversion of dihydroflavonols into flavonols and anthocyanins in flowers of Petunia hybrida. Planta 155: 364–368 (1982).
Hanahan D: Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166: 557–580 (1983).
Harker CL, Ellis THN, Coen ES: Identification and genetic regulation of the chalcone synthase multigene family in pea. Plant Cell 2: 185–194 (1990).
Helariutta Y, Elomaa P, Kotilainen M, Seppänen P, Teeri TH: Cloning of cDNA coding for dihydroflavonol-4-reductase (DFR) and characterization of dfr expression in the corollas of Gerbera hybrida var. Regina (Compositae). Plant Mol Biol 22: 183–193 (1993).
Hrazdina G, Kreuzaler F, Hahlbrock K, Grisebach H: Substrate specificity of flavanone synthase from cell suspension culture of parsley and structure of release products in vitro. Arch Biochem Biophys 169: 84–90 (1976).
Jackson D, Roberts K, Martin C: Temporal and spatial control of expression of anthocyanin biosynthetic genes in developing flowers of Antirrhinum majus. Plant J 2: 425–434 (1992).
Jones JDG, Duinsmuir P, Bedbrook J: High level expression of introduced chimeric genes in regenerated transformed plants. EMBO J 4: 2411–2418 (1985).
Koes RE, Spelt CE, Mol JNM: The chalcone synthase multigene family of Petunia hybrida (V30): differential, light-regulated expression during flower development and UV light induction. Plant Mol Biol 12: 213–225 (1989).
Koes RE, Spelt CE, van den Elzen PJM, Mol JNM: Cloning and molecular characterization of the chalcone synthase multigene family of Petunia hybrida. Gene 81: 245–257 (1989).
Koes RE, van Blokland R, Quattrocchio F, van Tunen AJ, Mol JNM: Chalcone synthase promoters in petunia are active in pigmented and unpigmented cell types. Plant Cell 2: 379–392 (1990).
Koes RE, Quattrocchio F, Mol JNM: The flavonoid biosynthetic pathway in plants: function and evolution. Bioessays 16: 123–132 (1994).
Kreuzaler F, Hahlbrock K: Enzymatic synthesis of aromatic compounds in higher plants. Formation of bis-noryangonin (4-hydroxy-6[4-hydroxystyryl]2-pyrone) from p-coumaroyl-CoA and malonyl-CoA. Arch Biochem Biophys 169: 84–90 (1975).
Lanz T, Tropf S, Marner F-J, Schröder J, Schröder G: The role of cysteines in polyketide synthases: site-directed mutagenesis of resveratrol and chalcone synthases, two key enzymes in different plant-specific pathways. J Biol Chem 266: 9971–9976 (1991).
Martin C, Carpenter R, Coen ES, Gerats AGM: The control of floral pigmentation in Antirrhinum majus. In: Thomas H, Grierson D (eds) Developmental Mutations in Plants (SEB Seminar Series 32), pp. 19–53. Cambridge University Press, Cambridge (1987).
Martin C, Gerats T: Control of pigment biosynthesis genes during petal development. Plant Cell 5: 1253–1264 (1993).
Melchior F, Kindl H: Grapevine stilbene synthase cDNA only slightly differing from chalcone synthase cDNA is expressed in Escherichia coli into catalytically active enzyme. FEBS Lett 268: 17–20 (1990).
Nakajima O, Akiyama T, Hakamatsuka T, Shibuya M, Noguchi H, Ebizuka Y, Sankawa U: Isolation, sequencing and bacterial expression of a cDNA for chalcone synthase from the cultured cells of Pueraria lobata. Chem Pharm Bull 39: 1911–1913 (1991).
Niesbach-Klösgen U, Barzen E, Bernhardt J, Rohde W, Schwarz-Sommer Z, Reif HJ, Wienand U, Saedler H: Chalcone synthase genes in plants: a tool to study evolutionary relationships. J Mol Evol 26: 213–225 (1987).
Ohl S, Hahlbrock K, Schäfer E: A stable blue-light-derived signal modulates ultraviolet-light-induced activation of the chalcone-synthase gene in cultured parsley cells. Planta 177: 228–236 (1989).
Proksch P, Rodriquez E: Chromenes and benzofurans of the Asteraceae. Their chemistry and biological significance. Phytochemistry 22: 2335–2348 (1983).
Quattrocchio F, Wing JF, Leppen HTC, Mol JNM, Koes RE: Regulatory genes controlling anthocyanin pigmentation are functionally conserved among plant species and have distinct sets of target genes. Plant Cell 5: 1497–1512 (1993).
Recourt K, van Tunen AJ, Mur L, van Brussel AAN, Lugtenberg BJJ, Kijne JW: Activation of flavonoid biosynthesis in roots of Vicia sativa subsp. nigra plants by inoculation with Rhizobium leguminosarum biovar viciae. Plant Mol Biol 19: 411–420 (1992).
Reimold U, Kroeger M, Kreuzaler F, Hahlbrock K: Coding and 3′ non-coding nucleotide sequence of chalcone synthase mRNA and assignment of amino acid sequence of the enzyme. EMBO J 2: 1801–1805 (1983).
Sambrook J, Fritsch EF, Maniatis T: Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York (1989).
Sancar A, Hack AM, Rupp WD: Simple method for identification of plasmid-coded proteins. J Bact 137: 692–693 (1979).
Schanz S, Schröder G, Schröder J: Stilbene synthase from Scots pine (Pinus sylvestris). FEBS Lett 313: 71–74 (1992).
Schmid J, Doerner PW, Clouse SD, Dixon RA, Lamb CJ: Developmental and environmental regulation of a bean chalcone synthase promoter in transgenic tobacco. Plant Cell 2: 619–631 (1990).
Schröder G, Brown JWS, Schröder J: Molecular analysis of resveratrol synthase: cDNA, genomic clones and relationship with chalcone synthase. Eur J Biochem 172: 161–169 (1988).
Schröder J, Schröder G: Stilbene and chalcone synthases: related enzymes with key functions in plant specific pathways. Z Naturforsch 45c: 1–8 (1990).
Schüz R, Heller W, Hahlbrock K: Substrate specificity of chalcone synthase from Petroselinum hortense. J Biol Chem 258: 6730–6734 (1983).
Shen JB, Hsu F: Brassica anther-specific genes: characterization and in situ localization of expression. Mol Gen Genet 234: 379–389 (1992).
Siebertz R, Proksch P, Witte L: Accumulation and biosynthesis of the chromenes precocene I and II in Ageratum houstonianum. Phytochemistry 29: 2135–2138 (1990).
Sommer H, Saedler H: Structure of the chalcone synthase gene of Antirrhinum majus. Mol Gen Genet 202: 429–434 (1986).
Sparvoli F, Martin C, Scienza A, Gavazzi G, Tonelli C: Cloning and molecular analysis of structural genes involved in flavonoid and stilbene biosynthesis in grape (Vitis vinifera L.). Plant Mol Biol 24: 743–755 (1994).
Stich K, Eidenberger T, Wurst F, Forkmann G: Flavonol synthase activity and regulation of flavonol and anthocyanin biosynthesis during flower development in Dianthus caryophyllus L. (carnation). Z Naturforsch 47c: 553–560 (1992).
Takkinen K, Laukkanen M-L, Sizmann D, Alfthan K, Immonen T, Vanne L, Kaartinen M, Knowles JKC, Teeri TT: An active single-chain antibody containing a cellulase linker domain is secreted by Escherichia coli. Protein Eng 4: 837–841 (1991).
Taylor LP, Briggs W: Genetic regulation and photocontrol of anthocyanin accumulation in maize seedlings. Plant Cell 2: 115–128 (1990).
Taylor LP, Jorgensen R: Conditional male fertility in chalcone synthase-deficient petunia. J Hered 83: 11–17 (1992).
Tropf S, Lanz T, Rensing SA, Schröder J, Schröder G: Evidence that stilbene synthases have developed from chalcone synthases several times in the course of evolution. J Mol Evol 38: 610–618 (1994).
van Brussel AAN, Recourt K, Pees E, Spaink HP, Teun T, Wijffelman CA, Kijne JW, Lugtenberg BJJ: A biovar-specific signal of Rhizobium leguminosarum bv. viciae induces increased nodulation gene-inducing activity in root exudate of Vicia sativa subsp. nigra. J Bact 172: 5394–5401 (1990).
van der Meer IM, Stam ME, van Tunen AJ, Mol JNM, Stuije AR: Antisense inhibition of flavonoid biosynthesis in petunia anthers results in male sterility. Plant Cell 4: 253–262 (1992).
Weiss D, van der Huit AH, Kroon JTM, Mol JNM, Kooter JM: The Petunia homologue of the Antirrhinum candi and Zea mays A2 flavonoid genes; homology to flavanone 3-hydroxylase and ethylene forming enzyme. Plant Mol Biol 22: 893–897 (1993).
Wienand U, Weydemann U, Niesbach-Klösgen U, Peterson PA, Saedler H: Molecular cloning of the c2 locus of Zea mays the gene coding for chalcone synthase. Mol Gen Genet 203: 202–207 (1986).
Wingender R, Röhrig H, Höriche C, Wing D, Schell J: Differential regulation of soybean chalcone synthase genes in plant defense, symbiosis and upon environmental stimuli. Mol Gen Genet 218: 315–322 (1989).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Helariutta, Y., Elomaa, P., Kotilainen, M. et al. Chalcone synthase-like genes active during corolla development are differentially expressed and encode enzymes with different catalytic properties in Gerbera hybrida (Asteraceae). Plant Mol Biol 28, 47–60 (1995). https://doi.org/10.1007/BF00042037
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00042037