Abstract
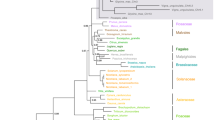
The comparative analysis of a large number of plant cyclins of the A/B family has recently revealed that plants possess two distinct B-type groups and three distinct A-type groups of cyclins [1]. Despite earlier uncertainties, this large-scale comparative analysis has allowed an unequivocal definition of plant cyclins into either A or B classes. We present here the most important results obtained in this study, and extend them to the case of plant D-type cyclins, in which three groups are identified. For each of the plant cyclin groups, consensus sequences have been established and a new, rational, plant-wide naming system is proposed in accordance with the guidelines of the Commission on Plant Gene Nomenclature. This nomenclature is based on the animal system indicating cyclin classes by an upper-case roman letter, and distinct groups within these classes by an arabic numeral suffix. The naming of plant cyclin classes is chosen to indicate homology to their closest animal class. The revised nomenclature of all described plant cyclins is presented, with their classification into groups CycA1, CycA2, CycA3, CycB1, CycB2, CycD1, CycD2 and CycD3.
Similar content being viewed by others
References
Renaudin JP, Savouré A, Philippe H, Van montagu M, Inzé D, Rouzé P, Characterization and classification of plant cyclins sequences related to A and B-type cyclins. In: Francis D, Dudits D, Inzé D (eds) Plant Cell Division, Portland Press, London, in press.
Pines J: Cyclins and cyclin-dependent kinases: a biochemical view. Biochem J 308: 697–711 (1995).
Nigg EA: Cyclin-dependent protein kinases: key regulators of the eukaryotic cell cycle. Bioessays 17: 471–480 (1995).
Norbury C, Nurse P: Animal cell cycles and their control. Annu Rev Biochem 61: 441–470 (1992).
Evans T, Rosenthal ET, Youngblom J, Distel D, Hunt T: Cyclin: A protein specified by maternal mRNA in sea urchin eggs that is destroyed at each cleavage division. Cell 33: 389–396 (1983).
Meijer L, Arion D, Golsteyn R, Pines J, Brizuela L, Hunt T, Beach D: Cyclin is a component of the sea urchin egg M-phase specific histone H1 kinase. EMBO J 8: 2275–2282 (1989).
Nakamura T, Sanokawa R, Sasaki YF, Ayusawa D, Oishi M, Mori N: Cyclin I: a new cyclin encoded by a gene isolated from human brain. Exp Cell Res 221: 534–542 (1995).
Pines J: Cyclins and cyclin-dependent kinases: Theme and variations. Adv Cancer Res 66: 181–212 (1995).
Fitch I, Dahmann C, Surana U, Amon A, Nasmyth K, Goetsch L, Byers B, Futcher B: Characterization of four B-type cyclin genes of the budding yeast Saccharomyces cerevisiae. Mol Biol Cell 3: 805–818 (1992).
Fisher D, Nurse P: Cyclins of the fission yeast Schizosaccharomyces pombe. Semin Cell Biol 6: 73–78 (1995).
Schwob E, Nasmyth K: CLB5 and CLB6, a new pair of B cyclins involved in DNA replication in Saccharomyces cerevisiae. Genes Devel. 7: 1160–1175 (1993).
Cross FR: Starting the cell cycle: What's the point? Curr Opin Cell Biol 7: 790–797 (1995).
Dirick L, Bohm T, Nasmyth K: Roles and regulation of Cln-Cdc28 kinases at the start of the cell cycle of Saccharomyces cerevisiae. EMBO J 14: 4803–4813 (1995).
Nugent JHA, Alfa CE, Young T, Hyams JS: Conserved structural motifs in cyclins identified by sequence analysis. J Cell Sci 99: 669–674 (1991).
O'Farrell P, Leopold P: A consensus of cyclin sequences reveals homology with the ras oncogene. Cold Spring Harbor Symp Quant Biol 56: 83–92 (1991).
Zheng XF, Ruderman JV: Functional analysis of the P Box, a domain in cyclin B required for the activation of Cdc25. Cell 75: 155–164 (1993).
Kobayashi H, Stewart E, Poon R, Adamczewski JP, Gannon J, Hunt T: Identification of the domains in cyclin A required for the binding to, and activation of, p34cdc2 and p32cdk2 protein kinase subunits. Mol Biol Cell 3: 1279–1294 (1992).
Lees EM, Harlow E: Sequences within the conserved cyclin box of human cyclin A are sufficient for binding to and activation of cdc2 kinase. Mol Cell Biol 13: 1194–1201 (1993).
Jeffrey PD, Russo AA, Polyak K, Gibbs E, Hurwitz J, Massagué J, Pavletich NP: Mechanism of cdk activation revealed by the structure of a cyclinA-CDK2 complex. Nature 376: 313–320 (1995).
Brown NR, Noble MEM, Endicott JA, Garman EF, Wakatsuki S, Mitchell E, Rasmussen B, Hunt T, Johnson LN: The crystal structure of cyclin A. Structure 3: 1235–1247 (1995).
Shaul O, Van montagu M, Inzé D: Regulation of cell division in Arabidopsis. Crit Rev Plant Sci 15: 97–112 (1996).
Jacobs T: Control of the cell cycle. Devel Biol 153: 1–15 (1992).
Jacobs TW: Cell cycle control. Annu Rev Plant Physiol Plant Mol Biol 46: 317–339 (1995).
Ferreira P, Hemerly A, Van Montagu M, Inzé D: Control of cell proliferation during plant development. Plant Mol Biol 26: 1289–1303 (1994).
Francis D, Halford NG: The plant cell cycle. Physiol Plant 93: 365–374 (1995).
Renaudin JP, Colasanti J, Rime H, Yuan Z, Sundaresan V: Cloning of four cyclins from maize indicates that higher plants have three structurally distinct groups of mitotic cyclins. Proc Natl Acad Sci USA 91: 7375–7379 (1994).
Kouchi H, Sekine M, Hata S: Distinct classes of mitotic cyclins are differentially expressed in the soybean shoot apex during the cell cycle. Plant Cell 7: 1143–1155 (1995).
Ferreira P, Hemerly A, Engler JD, Bergounioux C, Burssens S, Van Montagu M, Engler G, Inzé D: Three discrete classes of Arabidopsis cyclins are expressed during different intervals of the cell cycle. Proc Natl Acad Sci USA 91: 11313–11317 (1994).
Soni R, Carmichael JP, Shah ZH, Murray JAH: A family of cyclin D homologs from plants differentially controlled by growth regulators and containing the conserved retinoblastoma protein interaction motif. Plant Cell 7: 85–103 (1995).
Dahl M, Meskiene I, Bogre L, Ha DTC, Swoboda I, Hubmann R, Hirt H, Heberlebors E: The D-type alfalfa cyclin gene cycMs4 complements G(1) cyclin-deficient yeast and is induced in the G(1) phase of the cell cycle. Plant Cell 7: 1847–1857 (1995).
Lonsdale DM, Price CA: Eukaryotic gene nomenclature: a resolvable problem? Trends Biochem Sci, in press (1996).
Price CA, Reardon EM, Lonsdale DM: A guide to naming sequenced plant genes. Plant Mol Biol 30: 225–227 (1996).
Herendeen PS, Crane PR, The fossil history of the monocotyledons. In: Ruddall et al. (ed) Monocotyledons: Systematics and Evolution, pp. 1–21. Royal Botanical Gardens, Kew (1995).
Glotzer M, Murray AW, Kirschner MW: Cyclin is degraded by the ubiquitin pathway. Nature 349: 132–138 (1991).
Lorca T, Devault A, Colas P, VanLoon A, Fesquet D, Lazaro JB, Dorée M: Cyclin A-Cys41 does not undergo cell cycle-dependent degradation in Xenopus extracts. FEBS Lett 306: 90–93 (1992).
Pines J, Hunter T: The differential localization of human cyclins A and B is due to a cytoplasmic retention signal in cyclin B. EMBO J 13: 3772–3781 (1994).
Pines J, Hunter T: Human cyclins A and B1 are differentially located in the cells and undergo cell cycle dependent nuclear transport. J. Cell Biol 115: 1–17 (1991).
Ookata K, Hisanaga S, Bulinski JC, Murofushi H, Aizawa H, Itoh TJ, Hotani H, Okumura E, Tachibana K, Kishimoto T: Cyclin B interaction with microtubule-associated protein 4 (MAP4) targets p34(cdc2) kinase to microtubules and is a potential regulator of M-phase microtubule dynamics. J Cell Biol 128: 849–862 (1995).
Jackman M, Firth M, Pines J: Human cyclins B1 and B2 are localized to strikingly different structures: B1 to microtubules, B2 primarily to the Golgi apparatus. EMBO J 14: 1646–1654 (1995).
Furuya M: Cell division patterns in multicellular plants. Annu Rev Plant Physiol 35: 349–373 (1984).
Hata S, Kouchi H, Suzuka I, Ishii T: Isolation and characterization of cDNA clones for plant cyclins. EMBO J 10: 2681–2688 (1991).
Weinberg RA: The retinoblastoma protein and cell cycle control. Cell 81: 323–330 (1995).
Dowdy SF, Hinds PW, Louie K, Reed SI, Arnold A, Weinberg RA: Physical interaction of the retinoblastoma protein with human D cyclins. Cell 73: 499–511 (1993).
Rechsteiner M: Regulation of enzyme levels by proteolysis: the role of PEST regions. Adv Enzyme Regul 27: 135–151 (1988).
Rogers S, Wells R, Rechsteiner M: Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science 234: 364–368 (1986).
Gigot C, Spiker S: Nomenclature of genes encoding histones. Plant Mol Biol Rep 12: S39-S40 (1994).
Shaul O, Mironov V, Burssens S, Van montagu M, Inzé M: Two Arabidopsis cyclin promoters mediate distinctive transcriptional oscillation in synchronized tobacco BY-2 cells. Proc Natl Acad Sci USA 93: 4868–4872 (1996).
Setiady YY, Sekine M, Hariguchi N, Yamamoto T, Kouchi H, Shinmyo A: Tobacco mitotic cyclins: cloning, characterization, gene expression and functional assay. Plant J 8: 949–957 (1995).
Meskiene I, Bogre L, Dahl M, Pirck M, Ha DTC, Swoboda I, Heberlebors E, Ammerer G, Hirt H: cycMs3, a novel B-Type alfalfa cyclin gene, is induced in the G(0)-to-G(1) transition of the cell cycle. Plant Cell 7: 759–771 (1995).
Hirt H, Mink M, Bögre L, Györgyey J, Jonak C, Gartner A, Dudits D, Heberle-Bors E: Alfalfa cyclins: differential expression during the cell cycle and in plant organs. Plant Cell 4: 1531–1538 (1992).
Fobert PR, Coen ES, Murphy GJP, Doonan JH: Patterns of cell division revealed by transcriptional regulation of genes during the cell cycle in plants. EMBO J 13: 616–624 (1994).
Sauter M, Mekhedov SL, Kende H: Gibberellin promotes histone H1 kinase activity and the expression of cdc2 and cyclin genes during the induction of rapid growth in deepwater rice internodes. Plant J 7: 623–632 (1995).
Savouré A, Feher A, Kalo P, Petrovics G, Csanadi G, Szecsi J, Kiss G, Brown S, Kondorosi A, Kondorosi E: Isolation of a full-length mitotic cyclin cDNA clone CycIIIMs from Medicago sativa: chromosomal mapping and expression. Plant Mol Biol 27: 1059–1070 (1995).
Hemerly A, Bergounioux C, Van Montagu M, Inzé D, Ferreira P: Genes regulating the plant cell cycle: Isolation of a mitotic-like cyclin from Arabidopsis thaliana. Proc Natl Acad Sci USA 89: 3295–3299 (1992).
Fuerst RAUL, Soni R, Murray JAH, Lindsey K: Cyclin transcripts in synchronized Arabidopsis cells. Plant Physiol, in press (1996).
Szarka S, Fitch M, Schaerer S, Moloney M: Classification and expression of a family of cyclin gene homologues in Brassica napus. Plant Mol Biol 27: 263–275 (1995).
Day IS, Reddy ASN, Golovkin M: Isolation of a new mitotic-like cyclin from Arabidopsis: complementation of a yeast cyclin mutant with a plant cyclin. Plant Mol Biol 30: 565–575 (1996).
Deckert J, Jelenska J, Gwozdz EA, Legocki AB: The isolation of a lupine cDNA clone coding for a putative cyclin protein. Biochimie 78: 90–94 (1996).
Qin LX, Richard L, Perennes C, Gadal P, Bergounioux C: Identification of a cell cycle-related gene, cyclin, in Nicotiana tabacum (L). Plant Physiol 108: 425–426 (1995).
Logemann E, Wu SC, Schroder J, Schmelzer E, Somssich IE, Hahlbrock K: Gene activation by UV light, fungal elicitor or fungal infection in Petroselinum crispum is correlated with repression of cell cycle-related genes. Plant J 8: 865–876 (1995).
Reichheld JP, Chaubet N, Shen WH, Renaudin JP, Gigot C: Multiple A-type cyclins express sequentially during the cell cycle in Nicotiana tabacum BY2 cells. Proc Natl Acad Sci USA, in press (1996).
Murray JAH, Freeman D, Greenwood J, Huntley R, Makkerh J, Riou-Khamlichi C, Sorrell DA, Cockcroft C, Carmichael JP, Soni R, Shah ZH: Plant D cyclins and retinoblastoma (Rb) protein homologues. In: Francis D, Dudits D, Inzé D, (eds.), Plant Cell Division. Portland Press, London, in press (1996).
Xie Q, Sanz-Burgos AP, Hannon GJ, Gutiérrez C: Plant cells contain a novel member of the retinoblastoma family of growth regulatory proteins. EMBO J, 15: 4900–4908 (1996).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Renaudin, JP., Doonan, J.H., Freeman, D. et al. Plant cyclins: a unified nomenclature for plant A-, B- and D-type cyclins based on sequence organization. Plant Mol Biol 32, 1003–1018 (1996). https://doi.org/10.1007/BF00041384
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00041384