Abstract
The expression of the two Cu,ZZn superoxide dismutase (SOD) genes of tomato was followed in different organs and plant developmental stages at the transcript and enzymatic activity levels. The cDNA clones used as probes [28] code for the chloroplast Cu, Zn SOD (clone T1) and the cytosolic Cu, Zn SOD (clone P31).
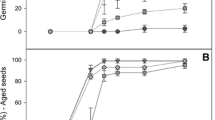
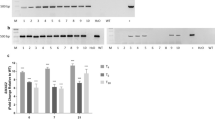
The two genes were found to display distinct expression patterns. While the T1 transcript was rare or absent from roots, stems and ripening fruits, the P31 transcript was very abundant in these organs. Shoot tips, flower buds, seedlings and young leaves contained high levels of the two mRNAs. During leaf expansion, the levels of both transcripts diminish markedly. Despite the diminished presence of transcripts, SOD activity levels of the corresponding cytosolic and chloroplast isozymes accumulated and were sustained throughout leaf expansion. In non-photosynthetic organs, the SOD-3 (cytosolic) isozyme contained most of the activity, while in the expanded leaf the SOD-1 (chloroplast) isozyme was more abundant. Light-regulated accumulation of both the P31 transcript (1.7-fold) and the T1 transcript (3-fold) was observed upon light exposure of etiolated seedlings. However, only SOD-1 activity was observed to increase, after a lag of a few hours.
The levels of both transcripts increased in response to paraquat and mechanical wounding. The level of the cytosolic transcript and the respective isozyme activity increased dramatically during prolonged drought stress while the chloroplast transcript remained unaffected. The expression of both genes was enhanced by spraying tomato plants with ethephon-a compound that releases ethylene.
Our data show that the expression of Cu, Zn SOD genes in tomato is modulated in response to a variety of factors and suggest the importance of oxyradical toxicity as well as the role of SOD in the defence mechanism of plants exposed to stress.
Similar content being viewed by others
References
Asada, K, Takahashi, M: Production and scavenging of active oxygen in photosynthesis. In: Kyle, DJ, Osmond, CV, Arntzen, CJ (eds) Photoinhibition, pp. 227–287, Elsevier, Amsterdam (1987).
Ausubel, FM, Brent, R, Kingston, RE, Moore, DD, Smith, H: Current Protocols in Molecular Biology. Green Brookly, NY (1990).
Baker, JE: Superoxide dismutase in ripening fruits. Plant Physiol 58: 644–647 (1976).
Baum, JA, Chandlee, JM, Scandalios, JG: Purification and partial characterization of a genetically defined superoxide dismutase (SOD-1) associated with maize chloroplasts. Plant Physiol 73: 31–35 (1983).
Baum, JA, Scandalios, JG: Developmental expression and intracellular localization of superoxide dismutase in maize. Differentiation 13: 133–140 (1979).
Baum, JA, Scandalios, JG: Expression of genetically distinet superoxide dismutases in the maize seedling during development. Devel Genet 3: 7–23 (1982).
Beauchamp, CO, Fridovich, I: Superoxide dismutase: improved assays and an assay applicable to acrylamide gel. Anal Biochem 44: 276–287 (1971).
Blech, DM, Borders, CL: Hydroperoxide anion, HO2, is an affinity reagent for the inactivation of yeast Cu, Zn superoxide dismutase: modification of one histidine per subunit. Arch Biochem Biophys 224: 579–586 (1983).
Burke, JJ, Gamble, PE, Hatfield, JL, Quisenberry, JE: Plant morphological and biochemical responses to field water deficit. I. Responses of glutathione reductase activity and paraquat sensitivity. Plant Physiol 79: 415–419 (1985).
Christman, MF, Morgan, RW, Jacobson, FS, Ames, BN: Positive control of a regulon for defense against oxidative stress and some heat-shock proteins in Salmonella typhimurium. Cell 41: 753–762 (1985).
Del, Rio, LA, Sevilla, F, Gomez, M, Yanez, J, Lopez, J: Superoxide dismutase: an enzyme system for the study of micronutrient interactions in plants. Planta 140: 221–225 (1978).
Dhindsa, RS, Plumb-Dhindsa, P, Thorpe, TA: Leaf senescence: correlated with increased levels of membrane permeability and lipid peroxidation, and decreased levels of superoxide dismucase and catalase. J Exp Bot 32: 93–101 (1981).
Elstner, EF: Oxygen activation and oxygen toxicity. Ann Rev Plant Physiol 33: 73–96 (1982).
Halliwell, B: The toxic effect of oxygen on plant tissues. In: Oberley, LW (ed) Superoxide Dismutase vol I, pp. 89–123, CRC Press, Florida (1982).
Harris, N, Dodge, AD: The effect of paraquat on flax cotyledon leaves: changes in fine structure. Planta 104: 201–209 (1972).
Hassan, HM: Microbial superoxide dismutases. In: Scandalios, JG (ed) Advances in Genetics, vol 26, pp. 65–97, Academic Press, New York. (1989).
Ishii, S: Generation of active oxygen species during enzymic isolation of protoplasts from oat leaves. In vitro Cell Devel Biol 23: 653–658 (1987).
Jansen, MAK, Malan, C, Shaaltiel, Y, Gressel, J: Mode of evolved photo oxidant resistance to herbicides and xenobiotics. Z Naturforsch 45c: 463–469 (1990).
Kwiatowski, J, Kaniuga, Z: Evidence for iron-containing superoxide dismutase in leaves of L. esculentum and Phaseolus vulgaris. Acta Physiol Plant 6: 197–202 (1984).
Kwiatowski, J, Kaniuga, Z: Isolation and characterization of cytosolic and chloroplastic isozymes of Cu, Zn superoxide dismutase from tomato leaves and their relationships to other Cu, Zn superoxide dismutases. Biochim Biophys Acta 874: 99–115 (1986).
Kwiatowski, J, Safianowska, A, Kaniuga, Z: Isolation and characterization of an iron-containing superoxide dismutase from tomato leaves, Lycopersicon esculentum. Eur J Biochem 146: 459–466 (1985).
Laemmli, AK: Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 (1970).
Logemann, J, Schell, J, Willmitzer, L: Improved method for the isolation of RNA from plant tissues. Anal Biochem 163: 16–20 (1987).
Malan, C, Greyling, MM, Gressel, J: Correlation between Cu, Zn superoxide dismutase and glutathione reductase and environmental and xenobiotic stress tolerance in maize inbreds. Plants Sci 69: 157–166 (1990).
Maniatis, T, Fritsch, EF, Sambrook, J: Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY (1982).
Matters, GL, Scandalios, JG: Effect of the free radical-generating herbicide paraquat on the expression of the superoxide dismutase (SOD) genes in maize. Biochim Biophys Acta 882: 29–38 (1986).
Monk, LS, Fagerstedt, KV, Crawford, RMM: Oxygen toxicity and superoxide dismutase as an antioxidant in physiological stress. Physiol Plant 76: 456–459 (1989).
Perl-Treves, R, Nacmias, B, Aviv, D, Zeelon, EP, Galun, E: Isolation of two cDNA clones from tomato containing two different superoxide dismutase sequences. Plant Mol Biol 11: 609–623 (1988).
Perl-Treves, R, Abu-Abied, M, Magal, N, Galun, E, Zamir, D: Genetic mapping of tomato cDNA clones encoding the chloroplastic and the cytosolic isozymes of superoxide dismutase. Biochem Gen 28: 543–552 (1990).
Rabinowitch, HD, Clare, DA, Crapo, JD, Fridovich, I: Positive correlation between SOD and resistance to paraquat toxicity in the green alga Chlorella sorokiniana. Arch Biochem Biophys 225: 640–648 (1983).
Rabinowitch, HD, Fridovich, I: Superoxide radicals, superoxide dismutase and oxygen toxicity in plants. Photochem Photobiol 37: 679–690 (1983).
Rabinowitch, HD, Sklan, D: Superoxide dismutase: a possible protective agent against sunscald in tomatoes (Lycopersicon esculentum Mill.) Planta 148: 162–167 (1980).
Salin, ML, Bridges, SM: Chemiluminescence in wounded root tissue. Evidence for peroxidase involvement. Plant Physiol 67: 43–46 (1981).
Sekizawa, Y, Haga, M, Hirabayashi, E, Takeuchi, N, Takino, Y: Dynamic behaviour of superoxide generation in rice leaf tissue infected with blast fungus and its regulation by some substances. Agric Biol Chem 51: 763–770 (1987).
Shaaltiel, Y, Gressel, J: Multienzyme oxygen radical detoxification system correlated with paraquat resistance in Conyza bonariensis. Pestic Biochem Physiol 26: 22–28 (1986).
Tanaka, K, Sugahara, K: Role of superoxide dismutase in defence against SO2 toxicity and an increase in superoxide dismutase activity with SO2 fumigation. Plant Cell Physiol 21: 601–611 (1980).
Tepperman, JM, Dunsmuir, P: Transformed plants with elevated levels of chloroplast superoxide dismutase are not more resistant to superoxide toxicity. Plant Mol Biol 14: 501–511 (1990).
Yang, SF: Biosynthesis and action of ethylene. Hort-Science 20: 41–45 (1985).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Perl-Treves, R., Galun, E. The tomato Cu,Zn superoxide dismutase genes are developmentally regulated and respond to light and stress. Plant Mol Biol 17, 745–760 (1991). https://doi.org/10.1007/BF00037058
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00037058