Abstract
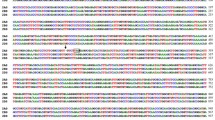
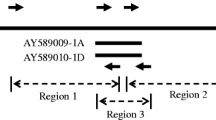
Myrosinase (thioglucoside glucohydrolase, EC 3.2.3.1.) is in Brassicaceae species such as Brassica napus and Sinapis alba encoded by two differentially expressed gene families, MA and MB, consisting of about 4 and 10 genes, respectively. Southern blot analysis showed that Arabidopsis thaliana contains three myrosinase genes. These genes were isolated from a genomic library and two of them, TGG1 and TGG2, were sequenced. They were found to be located in an inverted mode with their 3′ ends 4.4 kb apart. Their organization was highly conserved with 12 exons and 11 short introns. Comparison of nucleotide sequences of TGG1 and TGG2 exons revealed an overall 75% similarity. In contrast, the overall nucleotide sequence similarity in introns was only 42%. In intron 1 the unusual 5′ splice border GC was used. Phylogenetic analyses using both distance matrix and parsimony programs suggested that the Arabidopsis genes could not be grouped with either MA or MB genes. Consequently, these two gene families arose only after Arabidopsis had diverged from the other Brassicaceae species. In situ hybridization experiments showed that TGG1 and TGG2 expressing cells are present in leaf, sepal, petal, and gynoecium. In developing seeds, a few cells reacting with the TGG1 probe, but not with the TGG2 probe, were found indicating a partly different expression of these genes.
Similar content being viewed by others
References
Björkman R, Janson J-C: Studies on myrosinase I. Purification and characterization of a myrosinase from white mustard seed (Sinapis alba L.). Biochim Biophys Acta 276: 508–518 (1972).
Bones A, Iversen T-H: Myrosin cells and myrosinase. Isr J Bot 34: 351–376 (1985).
Chadchawan S, Bishop J, Thangstad OP, Bones AM, Mitchell-Olds T, Bradley D: Arabidopsis cDNA sequence encoding myrosinase. Plant Physiol 103: 671–672 (1993).
Drobnica L, Zemanova M, Nemec P, Antos K, Kristian P, Stullerova A. Knoppova V, Nemec PJr: Antifungal activity of isothiocyanate and related compounds. Appl Microbiol 15: 701–709 (1967).
Fahn A: Secretory Tissues in Plants. Academic Press, London (1979).
Falk A, Xue J, Lenman M, Rask L: Sequence of a cDNA clone encoding the enzyme myrosinase and expression of myrosinase in different tissues of Brassica napus. Plant Sci 83: 181–186 (1992).
Fenwick R, Heaney RK, Mullin WJ: Glucosinolates and their breakdown products in food and food plants. CRC Crit Rev Food Sci Nutr 18: 123–201 (1983).
Foster R, Izawa T, Chua N-H: Plant bZIP proteins gather at ACGT sites. FASEB J 8: 192–200 (1994).
Grange T, Roux J, Rigaud G, Pictet R: Cell-type specific activity of two glucocorticoid responsive units of rat tyrosine aminotransferase gene is associated with multiple binding sites for C/EBP and a novel liver-specific nuclear factor. Nucl Acids Res 19: 131–139 (1990).
Grob K, Matile P: Vacuolar location of glucosinolates in horseradish root cells. Plant Sci Lett 14: 327–335 (1979).
Haughn GW, Davin L, Giblin M, Underhill EW: Biochemical genetics of plant secondary metabolites in Arabidopsis thaliana. The glucosinolates. Plant Physiol 97: 217–226 (1991).
Higgins DG, Sharp PM: Fast and sensitive multiple sequence alignments on a microcomputer. Comput Appl Biosci 5: 151–153 (1989).
Holley RA, Jones JD: The role of myrosinase in the development of toxicity toward Nematospora in mustard seed. Can J Bot 63: 521–526 (1985).
Höglund A-S, Lenman M, Falk A, Rask L: Distribution of myrosinase in rapeseed tissues. Plant Physiol 95: 213–221 (1991).
Höglund A-S, Lenman M, Rask L: Myrosinase is localized to the interior of myrosin grains and is not associated to the surrounding tonoplast membrane. Plant Sci 85: 165–170 (1992).
James DC, Rossiter JT: Development and characterization of myrosinase in Brassica napus during early seedling growth. Physiol Plant 82: 163–170 (1991).
Kawagoe Y, Murai N: Four distinct nuclear proteins recognize in vitro the proximal promoter of the bean seed storage protein β-phaseolin gene conferring spatial and temporal control. Plant J 2: 927–936 (1992).
Kumar S, Tamura K, Nei M. MEGA: Molecular Evolutionary Genetics Analysis, version 1.01. The Pennsylvania State University, University Park, PA 16802 (1993).
Larsen PO, Glucosinolates. In: Stumpf PK, Tolbert NE, Conn EE (eds) The Biochemistry of Plants, vol 7. Secondary plant products, pp. 501–525. Academic Press, New York (1980).
Lenman M, Rödin J, Josefsson L-G, Rask L: Immunological characterization of rapeseed myrosinase. Eur J Biochem 194: 747–753 (1990).
Lenman M, Falk A, Xue J, Rask L: Characterization of a Brassica napus myrosinase pseudogene: myrosinases are members of the BGA family of β-glycosidases. Plant Mol Biol 21: 463–474 (1993).
Leman M, Falk A, Rödin J, Höglund A-S, Ek B, Rask L: Differential expression of myrosinase gene families. Plant Physiol 103: 703–711 (1993).
Louda SM, Rodman JE: Ecological patterns in the glucosinolate content of a native mustard. Cardamine cordifolia, in the Rocky Mountains. J Chem Ecol 9: 397–422 (1983).
Lönnerdal B, Janson J-C: Studies on myrosinase II. Purification and characterization of a myrosinase from rapeseed (Brassica napus L.). Biochim Biophys Acta 315: 421–429 (1973).
Machlin S, Mitchell-Olds T, Bradley D: Sequence of a Brassica campestris myrosinase gene. Plant Physiol 102: 1359–1360 (1993).
Prestridge DS: Signal scan: A computer program that scans DNA sequences for eukaryotic transcriptional elements. Comput Appl Biosci 7: 203–206 (1991).
Rodman JE: A taxonomic analysis of glucosinolate-producing plants. Part 1. Phenetics. Syst Bot 16: 598–618 (1991).
Saitou N, Nei M: The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 (1987).
Sang JP, Minchinton IR, Johnstone PK, Truscott JW: Glucosinolate profiles in the seed, root and the leaf tissue of cabbage, mustard, rapeseed, radish and swede. Can J Plant Sci 64: 77–93 (1984).
Srivastava VK, Hill DC: Glucosinolate hydrolytic products given by Sinapis alba and Brassica napus thioglucosidase. Phytochemistry 13: 1043–1046 (1974).
Staden R: An improved sequence handling package that runs on the Apple Machintosh. Comput Appl Biosci 4: 387–393 (1990).
Thangstad OP, Iversen TH, Slupphaug G, Bones A: Immunocytochemical localization of myrosinase in Brassica napus. Planta 180: 245–248 (1990).
Thangstad OP, Evjen K, Bones A: Immunogold-EM localization of myrosinase in Brassicaceae. Protoplasma 161: 85–93 (1991).
Thangstad OP, Winge P, Husebye H, Bones A: The myrosinase (thioglucoside glucohydrolase) gene family in Brassicaceae. Plant Mol Biol 23: 511–524 (1993).
Uppström B: Glucosinolate pattern in different growth stages of high and low glucosinolate varieties of Brassica napus. J Swed Seed Ass 4: 331–336 (1983).
Xue J, Lenman M, Falk A, Rask L: The glucosinolate-degrading enzyme myrosinase in Brassicaceae is encoded by a gene family. Plant Mol Biol 18: 387–398 (1992).
Xue J, Pihlgren U, Rask L: Temporal, cell-specific, and tissue-preferential expression of myrosinase genes during embryo and seedling development in Sinapis alba. Planta 191: 95–101 (1993).
Xue J, Rask L: The unusual 5′ splicing border GC is used in myrosinase genes of the Brassicaceae. Plant Mol Biol, submitted (1994).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Xue, J., Jørgensen, M., Pihlgren, U. et al. The myrosinase gene family in Arabidopsis thaliana: gene organization, expression and evolution. Plant Mol Biol 27, 911–922 (1995). https://doi.org/10.1007/BF00037019
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00037019