Abstract
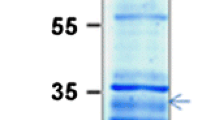
A cDNA clone for pre-ferredoxin-NADP+ reductase (FNR) was obtained by screening a Cyanophora paradoxa expression library with antibodies specific for cyanelle FNR. The 1.4 kb transcript was derived from a single-copy gene. The precursor (41 kDa) and mature forms (34 kDa) of FNR were identified by western blotting of in vitro translation products and cyanelle extracts, respectively. The derived amino acid sequence of the mature form was corroborated by data from N-terminal protein sequencing and yielded identity scores from 58% to 62% upon comparison with cyanobacterial FNRs. Sequence conservation seemed to be even more pronounced in comparison with enzymes from higher plants, but using the neighbor joining method the C. paradoxa sequence was clearly positioned between the prokaryotic and eukaryotic sequences. The transit peptide of 65 or 66 amino acids appeared to be totally unrelated to those from spinach, pea and ice plant but showed overall characteristics of stroma-targeting peptides.
Similar content being viewed by others
References
Aitken A, Stanier RY: Characterization of peptidoglycan from the cyanelles of Cyanophora paradoxa. J Gen Microbiol 212: 218–229 (1979).
Bayer MG: Detektion von kern- und cyanellenkodierten Proteinen bei Cyanophora paradoxa im Mikromaßstab sowie biochemische und molekularbiologische Charakterisierung des Ferredoxins. Thesis, University of Tübingen (1991).
Bayer MG, Gebhard UB, Maier TL, Schenk HEA: Two-step purification of Cyanophora ferredoxin and its identification in soluble protein preparations by isoelectric focusing. Protein Express Purif 2: 240–247 (1991).
Bayer MG, Maier TL, Gebhart UB, Schenk HEA: Cyanellar ferredoxin-NADP+-oxidoreductase is encoded by the nuclear genome and synthesized on cytoplasmic 80S ribosomes. Curr Genet 17: 265–267 (1990).
Bayer MG, Schenk HEA: Biosynthesis of proteins in Cyanophora paradoxa. I. Protein import into the endocyanelle analyzed by micro two-dimensional gel electrophoresis. Endocyt Cell Res 3: 197–202 (1986).
Bohnert HJ, Löffelhardt W: Molecular genetics of cyanelles from Cyanophora paradoxa In: Reisser W (ed) Algal Symbioses, pp. 379–397. Biopress, Bristol (1992).
Bryant DA: Genetic analysis of phycobilisome biosynthesis, assembly, structure, and function in the cyanobacterium Synechococcus sp. PCC 7002. In: Stevens SE, Bryant DA (eds) Light-Energy Transduction in Photosynthesis: Higher Plant and Bacterial Models, pp. 62–90. American Society of Plant Physiologists, Rockville (1988).
Ceccarelli EA, Viale AM, Krapp AR, Carrillo N: Expression, assembly, and processing of an active plant ferredoxin-NADP+ oxidoreductase and its precursor protein in Escherichia coli. J Biol Chem 266: 14283–14287 (1991).
Ellis RJ, van der Vies SM: Molecular chaperones. Annu Rev Biochem 60: 321–347 (1991).
Felsenstein J: Phylogenies from molecular sequences: Inference and reliability. Annu Rev Genet 22: 521–565 (1988).
Fillat MF, Bakker HAC, Weisbeek P: Sequence of the ferredoxin-NADP+ reductase from Anabaena PCC 7119. Nucl Acids Res 18: 7161 (1990).
Fitch WM, Margoliash E: Construction of phylogenetic trees. Science 155: 279–284 (1967).
Gebhart UB, Stefanovic S, Bayer MG, Maier TL, Schenk HEA: Ferredoxin-NADP+-oxidoreductase of Cyanophora paradoxa: purification, partial characterization, N-terminal amino acid sequence. Prot Express Purif 3: 228–235 (1992).
Hartl F-U, Neupert W: Protein sorting to mitochondria: evolutionary conservation of folding and assembly. Science 247: 930–938 (1990).
Harlow E, Lane D (eds) Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY (1988).
Henikoff S: Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene 28: 351–359 (1984).
Jansen T, Reiländer H, Stepphuhn J, Herrmann RG: Analysis of cDNA clones encoding the entire precursor-polypeptide for ferredoxin: NADP+ oxidoreductase from spinach. Curr Genet 13: 517–522 (1988).
Janssen I, Jakowitsch J, Michalowski C, Bohnert HJ, Löffelhardt W: Evolutionary relationship of psbA genes from cyanobacteria, cyanelles and plastids. Curr Genet 15: 335–340 (1989).
Karplus PA, Daniels MJ, Herriott JR: Atomic structure of ferredoxin-NADP+ reductase: prototype for a structurally novel flavoenzyme family. Science 251: 60–66 (1991).
Karplus PA, Walsh KA, Herriott JR: Amino acid sequence of spinach ferredoxin: NADP+ oxidoreductase. Biochemistry 23: 6576–6583 (1984).
Kraus M, Götz M, Löffelhardt W: The cyanelle str operon from Cyanophora paradoxa: Sequence analysis and phylogenetic implications. Plant Mol Biol 15: 561–573 (1990).
Kyte J, Doolittle RF: A simple method for displaying the hydropathic character of a protein. J Mol Biol 157: 105–132 (1982).
Lüttke A: MacPROT: a set of basic programs for protein structure analysis. Comp Meth Progr Biomed 31: 105–112 (1990).
Lüttke A: On the origin of chloroplasts and rhodoplasts: Protein sequence comparison. Endocyt Cell Res 8: 75–82 (1991).
Michalowski CB, Schmitt JM, Bohnert HJ: Expression during salt stress and nucleotide sequence of cDNA for ferredoxin-NADP+ reductase from Mesembryanthemum crystallinum. Plant Physiol 89: 817–822 (1989).
Neumann-Spallart C, Brandtner M, Kraus M, Jakowitsch J, Bayer MG, Maier TL, Schenk HEA, Löffelhardt W: The petFI gene encoding ferredoxin I is located close to the str operon on the cyanelle genome of Cyanophora paradoxa. FEBS Lett 268: 55–58 (1990).
Newman BJ, Gray JC: Characterisation of a full-length cDNA clone for pea ferredoxin-NADP+ reductase. Plant Mol Biol 10: 511–520 (1988).
Saitou N, Nei M: The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 (1987).
Schenk HEA, Bayer MG, Maier TL, Lüttke A, Gebhart UB, Stevanovic S: Ferredoxin-NADP+-oxidoreductase of Cyanophora paradoxa, nucleus-encoded but cyanobacterial. Gene transfer from symbiont to host, an evolutionary mechanism originating new species. Z Naturforsch 47c: 347–358 (1992).
Schenk HEA: Cyanobacterial symbioses. In: Ballows A, Trüper HG, Dworkin M, Harder W, Schleifer KH (eds) The Prokaryotes, vol. 4 pp. 3819–3854. Springer, New York (1992).
Schenk HEA: Nachweis einer lysozymempfindlichen Stütsmembran der Endocyanellen von Cyanophora paradoxa Korsch. Z Naturforsch 25b: 656 (1970).
Schluchter WM, Bryant DA: Molecular characterization of ferredoxin-NADP+ oxidoreductase in cyanobacteria: cloning and sequence of the petH gene of Synechococcus sp. PCC 7002 and studies on the gene product. Biochemistry 31: 3092–3102 (1992).
Shin M, Tsujita M, Tomizawa H, Sakihama N, Kamei K, Oshino R: Proteolytic degradation of ferredoxin-NADP+ reductase during purification from spinach. Arch Biochem Biophys 279: 97–103 (1990).
Short JM, Fernandez JM, Sorge JA, Huse WD: Lambda ZAP: a bacteriophage Lambda expression vector with in vivo excision properties. Nucl Acids Res 16: 7583–7600 (1988).
Smeekens S, Weisbeek P, Robinson C: Protein transport into and within chloroplasts. Trends Biochem Sci 15: 73–76 (1990).
Starnes SM, Lambert DH, Maxwell ES, Stevens SE, Porter RD, Shively JM: Cotranscription of the large and small subunit genes of ribulose-1.5-bisphosphate carboxylase/oxygenase in Cyanophora paradoxa. FEMS Microbiol Lett 28: 165–169 (1985).
von Heijne G: Chloroplast transit peptides: the perfect random coil? FEBS Lett 278: 1–3 (1991).
von Heijne G, Steppuhn J, Herrmann RG: Domain structure of mitochondrial and chloroplast targeting peptides. Eur J Biochem 180: 535–545 (1989).
Wang Y: Double-stranded DNA sequencing with T7 polymerase. Biotechniques 6: 843–845 (1988).
Wickner W, Driessen AJM, Hartl F-U: The enzymology of protein translocation across the Escherichia coli plasma membrane. Annu Rev Biochem 60: 101–124 (1991).
Yao Y, Tamura T, Wada K, Matsubara H, Kodo K: Spirulina ferredoxin-NADP+ reductase. The complete amino acid sequence. J Biochem 95: 1513–1516 (1984).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Jakowitsch, J., Bayer, M.G., Maier, T.L. et al. Sequence analysis of pre-ferredoxin-NADP+-reductase cDNA from Cyanophora paradoxa specifying a precursor for a nucleus-encoded cyanelle polypeptide. Plant Mol Biol 21, 1023–1033 (1993). https://doi.org/10.1007/BF00023600
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00023600