Abstract
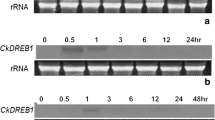
In this study we describe a novel gene, which was isolated in an attempt to search for specific plant resistance genes of Arabidopsis against isolates of the phytopathogenic bacterium Xanthomonas campestris pv. campestris. The gene was cloned by differential screening of a genomic library of the Xcc 750-resistant ecotype Col-0, using cDNA populations derived from ecotype Col-0 and the Xcc 750-susceptible ecotype Oy-0. The isolated gene, CXc750, is differentially expressed in ecotypes of Arabidopsis thaliana. In addition, although highly expressed in uninfected plants, gene expression increases in response to pathogen attack.
CXc750 potentially codes for a small, basic protein of about 10 kDa. The predicted protein product contains a potential signal leader peptide at the amino-terminal end but no ER retention sequence and no further transmembrane domain. This indicates that the gene product is transported to other compartments or out of the cell.
The possible function of CXc750 as a member of the plant defense response system is discussed.
Similar content being viewed by others
references
van den Ackerveken GF, van Kan JA, de Wit PG: Molecular evidence supporting the gene-for-gene hypothesis in Cladosporium fulvum-tomato interaction. Plant J 2: 359–366 (1992).
Ausubel FM: Current Protocols in Molecular Biology, Greene Publishing Associates/Wiley Interscience, New York (1989).
Bonas U, Stall RE, Staskawicz BJ: Genetic and structural characterization of the avirulence gene avr Bs3 from Xanthomonas campestris pv. vesicatoria. Mol Gen Genet 218: 127–136 (1989).
Bowles DJ: Defense-related proteins in higher plants. Annu Rev Biochem 59: 873–907 (1990).
Chomczynski P, Sacchi N: Single-step method of RNA isolation by acid guanidinium-thiocyanate phenol-chloroform extraction. Anal Biochem 102: 156–159 (1987).
Church GM, Gilbert W: Genomic sequencing. Proc Natl Acad Sci USA 81: 1991–1995 (1984).
Collinge DB, Slusarenko AJ: Plant gene expression in response to pathogens. Plant Mol Biol 9: 389–410 (1987).
Dangl JL, Lehnackers H, Kiedrowski S, Debener T, Rupprecht C, Arnold M, Somssich I. In: Hennecke H, Verma DPS (eds) Advances in Molecular Genetics of Plant-Microbe Interactions. Current Plant Science and Biotechnology in Agriculture vol. 1, pp. 78–83. Kluwer Academic Publishers, Dordrecht, Netherlands (1991).
Dangl JL, Ritter C, Gibbon MJ, Mur LAJ, Wood JR, Goss SG, Mansfield J, Taylor JD, Vivien A: Functional homologs of the Arabidopsis RPM1 disease resistance gene in bean and pea. Plant Cell 4: 1359–1369 (1992).
Daniels MJ, Fan MJ, Barber CE, Clarke BR, Parker JE: Interaction between Arabidopsis thaliana and Xanthomonas campestris. In: Hennecke H, Verma DPS (eds) Advances in Molecular Genetics of Plant — Microbe Interactions. Current Plant Science and Biotechnology vol. 1, pp. 84–89. Kluwer Academic Publishers, Dordrecht, Netherlands (1991).
Debener T, Lehnackers H, Arnold M, Dangl JL: Identification and molecular mapping of a single Arabidopsis thaliana locus determining resistance to a phytopathogenic Pseudomonas syringae isolate. Plant J 1: 289–302 (1991).
Dellaporta SL, Wood J, Hicks JB: A plant DNA minipreparation: version II. Plant Mol Biol Rep 1: 19–21 (1983).
Dixon RA: The phytoalexin response: elicitation, signalling and control of host gene expression. Biol Rev 61: 239–291 (1986).
Dixon RA, Harrison MJ: Activation, structure and organization of genes involved in microbial defense in plants. Adv Genet 28: 165–234 (1990).
Dixon RA, Lamb CJ: Molecular communication in interactions between plants and microbial pathogens. Annu Rev Plant Physiol Plant Mol Biol 41: 339–367 (1990).
Dong X, Mindrinos M, Davis KR, Ausubel FM: Induction of Arabidopsis defense genes by virulent and avirulent Pseudomonas syringae strains and by a cloned avirulence gene. Plant Cell 3: 61–72 (1991).
Ellingboe AH: Changing concepts in host-pathogen genetics. Annu Rev Phytopath 19: 125–143 (1981).
Feinberg AP, Vogelstein B: A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem 132: 6–13 (1983).
Flor H: Current status of gene-for-gene concept. Annu Rev Phytopath 9: 275–296 (1971).
Gabriel DW, Rolfe BG: Working models of specific recognition in plant-microbe interactions. Annu Rev Phytopath 28: 365–391 (1990).
Gurr SJ, McPherson MJC: PCR-directed cDNA libraries. In: McPherson MJC, Quirke P, Taylor GR (eds) PCR: A Practical Approach, pp. 147–171. IRL Press at Oxford University Press, London (1991).
Hahn MG, Bucheli P, Cervone F, Doares SH, O'Neill R et al. Roles of cell wall constituents in plant-pathogen interactions. In: Kosuge T, Nester EW (eds) Plant-Microbe Interactions: Molecular and Genetic Perspectives, pp. 131–181 (1989).
van Heijne G: A new method for predicting signal sequence cleavage sites. Nucl Acids Res 14: 4683–4690 (1986).
Jakobek JL, Lindgren PB: Generalized induction of defense responses in bean is not correlated with the induction of the hypersensitive reaction. Plant Cell 5: 49–56 (1993).
Joshi CP: Putative polyadenylation signals in nuclear genes of higher plants: A compilation and analysis. Nucl Acids Res 15: 9627–9640 (1987).
van Kan JAL, van den Ackerveken GF, de Wit PJ: Cloning and characterization of cDNA of avirulence gene avr 9 of the fungal pathogen Cladosporium fulvum causal agant of tomato leaf mold. Mol Plant-Microbe Interact 4: 52–59 (1991).
Keen NT, Staskawicz BJ: Host range determinants in plant pathogens and symbionts. Annu Rev Microbiol 42: 421–440 (1988).
Keen NT: Gene-for-gene complementarity in plant-pathogen interactions. Annu Rev Genet 24: 447–463 (1990).
Kiedrowski S, Kawalleck P, Hahlbrock K, Somssich I, Dangl JL: Rapid activation of a novel plant defense gene is strictly dependent on the Arabidopsis RPM1 disease resistance. EMBO J 11: 4677–4684 (1992).
Knogge W, Kombrink E, Schmelzer E, Hahlbrock K: Occurrence of phytoalexins and other putative defense-related substances in uninfected parsley plants. Planta 171: 279–287 (1987).
Koch E, Slusarenko AJ: Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell 2: 437–445 (1990).
Koncz C, Meyerhofer R, Koncz Kalman Z, Nawrath C, Reiss B, Redei GP, Schell J: Isolation of a gene encoding a novel chloroplast protein by T-DNA tagging in Arabidopsis thaliana. EMBO J 9: 1337–1346 (1990).
Lamb CJ, Lawton MA, Dron M, Dixon RA: Signals and transduction mechanisms for activation of plant defenses against microbial attack. Cell 56: 215–224 (1989).
Lawton MA, Lamb CJ: Transcriptional activation of plant defense genes by fungal elicitor, wounding and infection. Mol Cell Biol 7: 335–341 (1987).
Mahé A, Grisvard J, Desnos T, Dron M: Bean-Colletotrichum lindemuthianum compatible interactions: time course of plant defense responses depends on race. Mol Plant-Microbe Interact 4: 472–478 (1992).
Maniatis T, Fritsch EF, Sambrook J: Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY (1982).
Marton L, Browse J: Facile transformation of Arabidopsis. Plant Cell Rep 10: 235–239 (1991).
Michelmore RW, Kesseli RV, Francis DM, Fortin MG, Paran I, Chang-Hsien Y: Strategies for cloning plant disease resistance genes. In: Gurr SJ, McPherson MJ, Bowles DJ (eds) Molecular Plant Pathology. A Practical Approach, vol. 2, pp. 233–283 (1992).
Mogan BD, MacDonald MH, Graybosch R, Hunt AG: Upstream sequences other than AAUAAA are required for efficient mRNA 3′-end formation in plants. Plant Cell 2: 1261–1272 (1990).
Munro S, Pelham HRB: A C-terminal signal prevents secretion of luminal ER proteins. Cell 48: 899–907 (1987).
Sambrook J, Frisch EF, Maniatis T: Molecular Cloning: A Laboratory Manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY (1989).
Sanger F, Nicklen S, Coulson AR: DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA 74: 5463–5467 (1977).
Showalter AM: Structure and function of plant cell wall proteins. Plant Cell 5: 9–23 (1993).
Simpson RB, Johnson LJ: Arabidopsis thaliana as a host for Xanthomonas campestris pv. campestris. Mol Plant-Microbe Interact 3: 233–237 (1990).
Staskawicz BJ, Dahlbeck D, Keen N: Cloned avirulence gene of Pseudomonas syringae pv. glycinea determines race-specific incompatibility on Glycine max (L.) Merr. Proc Natl Acad Sci USA 81: 6024–6028 (1984).
Tsuji J, Somerville SC, Hammerschmidt R: Identification of a gene in Arabidopsis thaliana that controls resistance to Xanthomonas campestris pv. campestris. Physiol Mol Plant Path 38: 57–65 (1991).
Whalen MC, Innes RW, Bent AF, Staskawicz BJ: Identification of Pseudomonas syringae pathogens of Arabidopsis and a bacterial locus determining avirulence on both Arabidopsis and soybean. Plant Cell 3: 49–59 (1991).
de Wit PJG: Molecular characterization of gene-for-gene systems in plant-fungus interactions and the application of avirulence genes in control of plant pathogens. Annu Rev Phytopath 30: 391–418 (1992).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Aufsatz, W., Grimm, C. A new, pathogen-inducible gene of Arabidopsis is expressed in an ecotype-specific manner. Plant Mol Biol 25, 229–239 (1994). https://doi.org/10.1007/BF00023240
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00023240