Abstract
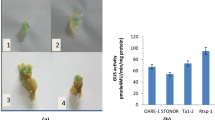
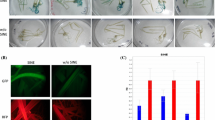
The BARE-1 retrotransposon occurs in more than 104 copies in the barley genome. The element is bounded by long terminal repeats (LTRs, 1829 bp) containing motifs typical of retrotransposon promoters. These, the presence of predicted priming sites for reverse transcription, and the high conservation for all key functional domains of the coding region suggest that copies within the genome could be active retrotransposons. In view of this, we looked for transcription of BARE-1 within barley tissues and examined the promoter function of the BARE-1 LTR. We demonstrate here that BARE-1-like elements are transcribed in barley tissues, and that the transcripts begin within the BARE-1 LTR downstream of TATA boxes. The LTR can drive expression of reporter genes in transiently transformed barley protoplasts. This is dependent on the presence of a TATA box functional in planta as well. Furthermore, we identify regions within the LTR responsible for expression within protoplasts by deletion analyses of LTR-luc constructs. Similarities between promoter regulatory motifs and regions of the LTR were identified by comparisons to sequence libraries. The activity of the LTR as a promoter, combined with the abundance of BARE-1 in the genome, suggests that BARE-1 may retain the potential for propagation in the barley genome.
Similar content being viewed by others
References
Allan AC, Trewavas AJ: Abscisic acid and gibberellin perception: inside or out? Plant Physiol 104: 1107–1108 (1994).
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ: Basic local alignment search tool. J Mol Biol 215: 403–410 (1990).
Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, Albright LM, Coen DM, Varki A, Janssen K: Current Protocols in Molecular Biology, pp. 4.7.1–4.7.8. John Wiley, New York (1995).
Beato M: Gene regulation by steroid hormones. Cell 56: 335–344 (1989).
Boeke JD, Corces VG: Transcription and reverse transcription of retrotransposons. Annu Rev Microbiol 43: 403–434 (1989).
Casacuberta JM, Grandbastien M-A: Characterization of LTR sequences involved in the protoplast specific expression of the tobacco Tnt1 retrotransposon. Nucl Acids Res 21: 2087–2093 (1993).
Chen G, Müller M, Potrykus I, Hohn T, Fütterer J: Rice tungro bacilliform virus: transcription and translation in protoplasts. Virology 204: 91–100 (1994).
Conklin KF: Activation of an endogenous retrovirus enhancer by insertion into a heterologous context. J Virol 65: 2525–2532 (1991).
Cullen BR, Greene WC: Regulatory pathways governing HIV-1 replication. Cell 58: 423–426 (1989).
Dinesh-Kumar SP, Miller WA: Control of start codon choice on a plant viral RNA encoding overlapping genes. Plant Cell 5: 679–692 (1993).
Donzé O, Damay P, Spahr P-F: The first and third uORFs in RSV leader RNA are efficiently translated: Implications for translational regulation and viral RNA packaging. Nucl Acids Res 23: 861–868 (1995).
Flavell AJ, Smith DR, Kumar A: Extreme heterogeneity of Ty1-copia group retrotransposons in plants. Mol Gen Genet 231: 233–242 (1992).
Hirochika H: Activation of tobacco retrotransposons during tissue culture. EMBO J 12: 2521–2528 (1993).
Hirochika H, Fukuchi A, Kikuchi F: Retrotransposon families in rice. Mol Gen Genet 233: 209–216 (1992).
Huang N, Sutliff TD, Litts JC, Rodriguez RL: Classification and characterization of the rice α-amylase mutigene family. Plant Mol Biol 14: 655–668 (1990).
Hurst HC, Parker MG: Rat prostatic steroid binding protein: DNA sequence and transcript maps of the two C3 genes. EMBO J 2: 769–774 (1983).
Huttly AK, Phillips AL, Tregear JW: Localization of cis elements in the promoter of a wheat α-Amy2 gene. Plant Mol Biol 19: 903–911 (1992).
Jin Y-K, Bennetzen JL: Structure and coding properties of Bs1, a maize retrovirus-like retrotransposon. Proc Natl Acad Sci USA 86: 6235–6239 (1989).
Joshi CP: An inspection of the domain between putative TATA box and translation start site in 79 plant genes. Nucl Acids Res 15: 6643–6653 (1987).
Joshi CP, Nuyen HT: 5′ untranslated leader sequences of eukaryotic mRNAs encoding heat shock induced proteins. Nucl Acids Res 23: 541–549 (1995).
Katz RA, Skalka AM: Generation of diversity in retroviruses. Annu Rev Genet 24: 409–445 (1990).
Keller B, Sauer N, Lamb CJ: Glycine-rich cell wall proteins in bean: gene structure and association of the protein with the vascular system. EMBO J 7: 3625–3633 (1988).
Kim J-K, Cao J, Wu R: Regulation and Interaction of multiple protein factors with the proximal promoter regions of a rice high PI α-amylase gene. Mol Gen Genet 232: 383–393 (1992).
Klaver B, Berkhout B: Comparison of 5′ and 3′ long terminal repeat promoter function in human immunodeficiency virus. J Virol 68: 3830–3840 (1994).
Konieczny A, Voytas DF, Cummings MP, Ausubel FM: A superfamily of Arabidopsis thaliana retrotransposons. Genetics 127: 801–809 (1991).
Kozak M: Structural features in eukaryotic mRNAs that modulate the initiation of translation. J Biol Chem 266: 19867–19870 (1991).
Lanahan MB, Ho T-HD, Rogers SW, Rogers JC: A gibberellin response complex in cereal α-amylase gene promoters. Plant Cell 4: 203–211 (1992).
Lee D, Ellis THN, Turner L, Hellens RP, Cleary WG: A copia-like element in Pisum demonstrates the use of dispersed repeated sequences in genetic analysis. Plant Mol Biol 15: 707–722 (1990).
Lucas H, Feuerbach F, Kunert K, Grandbastien M-A, Caboche M: RNA-mediated transposition of the tobacco retrotransposon Tnt1 in Arabidopsis thaliana. EMBO J 14: 2364–2373 (1995).
Luehrsen KR, de Wet JR, Walbot V: Transient expression analysis in plants using firefly luciferase reporter gene. Meth Enzymol 216: 397–414 (1992).
Manninen I, Schulman AH: BARE-1, a copia-like retroelement in barley (Hordeum vulgare L.). Plant Mol Biol 22: 829–846 (1993).
Melton DA, Krieg PA, Rebagliati MR, Maniatis T, Zinn K, Green MR: Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucl Acids Res 12: 7035–7056 (1984).
Moore G, Lucas H, Batty N, Flavell R: A family of retrotransposons and associated genomic variation in wheat. Genomics 10: 461–468 (1991).
Mundy J, Yamaguchi-Shinozaki K, Chua N-H: Nuclear proteins bind conserved elements in the abscisic acid-responsive promoter of a rice rab gene. Proc Natl Acad Sci USA 87: 1406–1410 (1990).
Oeda K, Salinas J, Chua N-H: A tobacco bZip transcription activator (TAF-1) binds to a G-box-like motif in plant genes. EMBO J 10: 1793–1802 (1991).
O'Neal JK, Pokalsky AR, Kiehne KL, Shewmaker CK: Isolation of tobacco SSU genes: Characterization of a transcriptionally active pseudogene. Nucl Acids Res 15: 8661–8677 (1987).
Pauls PK, Kunert K, Huttner E, Grandbastien M-A: Expression of the tobacco Tnt1 retrotransposon in heterologous species. Plant Mol Biol 26: 393–402 (1994).
Pouteau S, Huttner E, Grandbastien M-A, Caboche M: Specific expression of the tobacco Tnt1 retrotransposon in protoplasts. EMBO J 10: 1911–1918 (1991).
Renne R, Mergia A, Renshaw-Gegg LW, Neumann-Haefelin D, Luciw PA: Regulatory elements in the long terminal repeat (LTR) of Simian Foamy Virus Type 3 (SFV-3). Virology 192: 365–369 (1993).
Ritala A, Mannonen L, Aspegren K, Salmenkallio-Marttila M, Kurtén U, Hannus R, Mendez Lozano J, Teeri TH, Kauppinen V: Stable transformation of barley tissue culture by particle bombardment. Plant Cell Rep 12: 435–440 (1993).
Rogers JC, Lanahan MB, Rogers SW: The cis-acting gibberellin response complex in high-pI α-amylase gene promoters. Plant Physiol 105: 151–158 (1994).
Runeberg-Roos P, Kervinen J, Kovaleva V, Raikhel N, Gal S: The aspartic proteinase of barley is a vacuolar enzyme that processes probarley lectin in vitro. Plant Physiol 105: 321–329 (1994).
Rushton PJ, Hooley R, Lazarus CM: Aleurone nuclear proteins bind to similar elements in the promoter regions of two gibberellin-regulated α-amylase genes. Plant Mol Biol 19: 891–901 (1992).
Salmenkallio-Marttila M, Kauppinen V: Efficient regeneration of fertile plants from protoplasts isolated from microspore cultures of barley (Hordeum vulgare L.). Plant Cell Rep 14: 253–256 (1995).
Sambrook J, Fritsch EF, Maniatis T: Molecular Cloning: A Laboratory Manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY (1989).
Sandal NN, Bojsen K, Marcker K: A small family of nodule specific genes from soybean. Nucl Acids Res 15: 1507–1519 (1987).
Schmidt D, Graner A: Genomic organization and sequence diversity of the long terminal repeat (LTR) of the BARE-1 retrotransposon family in barley. Barley Genet Newsl 24: 24–27 (1994).
Shen Q, Ho T-HD: Functional dissection of an abscisic acid (ABA)-inducible gene reveals two independent ABA-responsive complexes each containing a G-box and a novel cis-acting element. Plant Cell 7: 295–307 (1995).
Skriver K, Olsen FL, Rogers JC, Mundy J: Cis-acting DNA elements responsive to gibberellin and its antagonist abscisic acid. Proc Natl Acad Sci USA 88: 7266–7270 (1991).
Smyth DR, Kalitsis P, Joseph JL, Sentry JW: Plant retrotansposon from Lilium henryi is related to Ty3 of yeast and the gypsy group of Drosophila. Proc Natl Acad Sci USA 86: 5015–5019 (1989).
Sparger EE, Shacklett BL, Renshaw-Gegg L, Barry PA, Pedersen NC, Elder JH, Luciw PA: Regulation of gene expression directed by the long terminal repeat of the feline immunodeficiency virus. Virology 187: 165–177 (1992).
Suoniemi A, Anamthawat-Jónsson K, Arna T, Schulman AH: Retrotransposon BARE-1 Is a major, dispersed component of the barley (Hordeum vulgare L.) genome. Plant Mol Biol (in press).
Sutliff TD, Lanahan MB, Ho T-HD: Gibberellin treatment stimulates nuclear factor binding to the gibberellin response complex in a barley α-amylase promoter. Plant Cell 5: 1681–1692 (1993).
Tada Y, Sakamoto M, Fujimura T: Efficient gene introduction into rice by electroporation and analysis of transgenic plants: use of electroporation buffer lacking chloride ions. Theor Appl Genet 80: 475–480 (1990).
Vander Wiel PL, Voytas DF, Wendel JF: Copia-like retrotransposable element evolution in diploid and polyploid cotton (Gossypium L.). J Mol Evol 36: 429–447 (1993).
Varmus HE: Retroviruses. In: Shapiro JA (ed) Mobile Genetic Elements, pp. 411–503. Academic Press, New York, (1983).
Voytas DF, Cummings MP, Konieczny AK, Ausubel FM, Rodermel SR: Copia-like retrotransposons are ubiquitous among plants. Proc Natl Acad Sci USA 89: 7124–7128 (1992).
Watanabe-Nagasu N, Itoh Y, Tani T, Okano K, Koga N, Okada N, Ohshima Y: Structural analysis of gene loci for Rat U1 small nuclear RNA. Nucl Acids Res 11: 1791–1801 (1983).
White SE, Habera LF, Wessler SR: Retrotransposons in the flanking regions of normal plant genes: A role for copia-like elements in the evolution of gene structure and expression. Proc Natl Acad Sci USA 91: 11792–11796 (1994).
Whittier RF, Dean DA, Rogers JC: Nucleotide sequence of alpha-amylase and thiole protease genes that are hormonally regulated in barley aleurone cells. Nucl Acids Res 15: 2515–2535 (1987).
Yamaguchi-Shinozaki K, Mundy J, Chua N-H: Four tightly linked rab genes are differentially expressed in rice. Plant Mol Biol 14: 29–39 (1989).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Suoniemi, A., Narvanto, A. & Schulman, A.H. The BARE-1 retrotransposon is transcribed in barley from an LTR promoter active in transient assays. Plant Mol Biol 31, 295–306 (1996). https://doi.org/10.1007/BF00021791
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00021791