Abstract
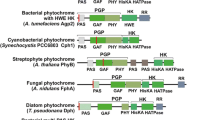
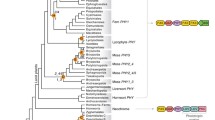
The phytochrome photoreceptor in the green alga Mesotaenium caldariorum is encoded by a small family of highly related genes. DNA sequence analysis of two of the algal phytochrome genes indicates an atypical gene structure with numerous long introns. The two genes, termed mesphy1a and mesphy1b, encode polypeptides which differ by one amino acid in the region of overlap that was sequenced. RT-PCR studies have established the intron-exon junctions of both genes and show that both are expressed. RNA blot analysis indicates a single transcript of ca. 4.1 kb in length. The deduced amino acid sequence of the mesphy1b gene reveals that the photoreceptor consists of 1142 amino acids, with an overall structure similar to other phytochromes. Phylogenetic analyses indicate that the algal phytochrome falls into a distinct subfamily with other lower plant phytochromes. Profile analysis of an internal repeat found within the central hinge region of the phytochrome polypeptide indicates an evolutionary relatedness to the photoactive yellow protein from the purple bacterium Ectothiorhodospira halophila, to several bacterial sensor kinase family members, and to a family of eukaryotic regulatory proteins which includes the period clock (per) and single-minded (sim) gene products of Drosophila. Since mutations which alter phytochrome activity cluster within the region delimited by these direct repeats (P.H. Quail et al., Science 268 (1995): 675–680), this conserved motif may play an important role in the signal transducing function of these disparate protein families.
Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ: Basic local alignment search tool. J Mol Biol 215: 403–410 (1990).
Antonsson C, Arulampalam V, Whitelaw ML, Pettersson S, Poellinger L: Constitutive function of the basic helix-loop-helix PAS factor arnt: regulation of target promoters via the E box motif. J Biol Chem 270: 13968–13972 (1995).
Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K: Current Protocols in Molecular Biology. Greene Publishing Associates/Wiley-Interscience, New York (1991).
Baca M, Borgstahl GEO, Boissinot M, Burke PM, Williams DR, Slater KA, Getzoff ED: Complete chemical structure of photoactive yellow protein: novel thioester-linked 4-hydroxycinnamyl chromophore and photocycle chemist. Biochemistry 33: 14369–14377 (1994).
Bassler BL, Wright M, Silverman MR: Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol Microbiol 13: 273–286 (1994).
Baylies MK, Vosshall LB, Sehgal A, Young MW: New short period mutations of the Drosophila clock gene Per. Neuron 9: 575–581 (1992).
Bendich AJ, Anderson RS, Ward BL: Plant DNA: long, pure and simple. In: Leaver CJ (ed) Genome Organization and Expression in Plants, pp. 31–33. Plenum Press, New York (1980).
Berkelman T, Lagarias JC: Calcium transport in the green alga Mesotaenium caldariorum: preliminary characterization and subcellular distribution. Plant Physiol 93: 748–757 (1990).
Blanco G, Drummond M, Woodley P, Kennedy C: Sequence and molecular analysis of the nifL gene of Azotobacter vinelandii. Mol Microbiol 9: 869–879 (1993).
Borgstahl GEO, Williams DR, Getzoff ED: 1.4 Å structure of photoactive yellow protein, a cytosolic photoreceptor: unusual fold, active site and chromophore. Biochemistry 34: 6278–6287 (1995).
Campbell WH, Gowri G: Codon usage in higher plants, green algae, and cyanobacteria. Plant Physiol 92: 1–11 (1990).
Canellakis ES, Paterakis AA, Huang SC, Panagiotidis CA, Kyriakidis DA: Identification, cloning, and nucleotide sequencing of the ornithine decarboxylase antizyme gene of Escherichia coli. Proc Natl Acad Sci USA 90: 7129–7133 (1993).
Chang C, Kwok SF, Bleecker AB, Meyerowitz EM: Arabidopsis ethylene-response gene Etr1: similarity of product to 2-component regulators. Science 262: 539–544 (1993).
Clack T, Mathews S, Sharrock RA: The phytochrome apoprotein family in Arabidopsis is encoded by five genes: the sequences and expression of PHYD and PHYE. Plant Mol Biol 25: 413–427 (1994).
Cowl JS, Hartley N, Xie DX, Whitelam GC, Murphy GP, Harberd NP: The phyC gene of Arabidopsis: absence of the third intron found in phyA and phyB. Plant Physiol 106: 813–814 (1994).
Crews ST, Thomas JB, Goodman CS: The Drosophila single-minded gene encodes a nuclear protein with sequence similarity to the Per gene product. Cell 52: 143–151 (1988).
Felsenstein J: PHYLIP: Phylogeny Inference Package. Department of Genetics, University of Washington, Seattle (1995).
Fischer RL, Goldberg RB: Structure and flanking regions of soybean seed protein genes. Cell 29: 651–660 (1982).
Furuya M: Phytochromes: their molecular species, gene families, and functions. Annu Rev Plant Physiol Plant Mol Biol 44: 617–645 (1993).
Garcher J: SeqVu Version 1.0. Garvan Institute of Medical Research (1995).
Goodall GJ, Filipowicz W: The AU-rich sequences present in the introns of plant nuclear pre-messenger RNAs are required for splicing. Cell 58: 473–483 (1989).
Gribskov M, Luthy R, Eisenberg D: Protile analysis. Meth Enzymol 183: 146–159 (1990).
Grolig F, Wagner G: Light-dependent chloroplast reorientations in Mougeotia and Mesotaenium: biased by pigment-regulated plasmalemma anchorage sites to actin filaments? Bot Acta 101: 2–6. (1987).
Hoffman EC, Reyes H, Chu FF, Sander F, Conley LH, Brooks BA, Hankinson O: Cloning of a factor required for activity of the Ah (dioxin) receptor. Science 252: 954–958 (1991).
Iuchi S, Fujiwara T, Lin ECC: The Arcb gene of Escherichia coli encodes a sensor-regulator protein for anaerobic repression of the Arc modulon. Mol Microbiol 4: 715–727 (1990).
Jayaratne P, Keenleyside WJ, Maclachlan PR, Dodgson C, Whitfield C: Characterization of Rcsb and Rcsc from Escherichia coli 09-K30 H12 and examination of the role of the Rcs regulatory system in expression of Group-I capsular polysaccharides. J Bact 175: 5384–5394 (1993).
Jones AM, Edgerton MD: The anatomy of phytochrome, a unique photoreceptor in plants. Sem Cell Biol 5: 295–302 (1994).
Kendrick RE, Kronenberg GHM: Photomorphogenesis in Plants. Martinus Nijhoff Publishers, Dordrecht, Netherlands (1994).
Kidd DG, Lagarias JC: Phytochrome from the green alga Mesotaenium caldariorum: purification and preliminary characterization. J Biol Chem 265: 7029–7035 (1990).
Kohrer K, Domdey H: Preparation of high molecular weight RNA. Meth Enzymol 194: 398–405 (1991).
Leong D, Boyer H, Betlach M: Characterization of a second gene involved in bacterio-opsin gene expression in a halophilic archaebacterium. J Bact 170: 4910–4915 (1988).
Lindebro MC, Poellinger L, Whitelaw ML: Protein-protein interaction via Pas domains: role of the Pas domain in positive and negative regulation of the Bhlh/Pas dioxin receptor-Arnt transcription factor complex. EMBO J 14: 3528–3539 (1995).
Luehrsen KR, Walbot V: Intron creation and polyadenylation in maize are directed by AU-rich RNA. Genes Devel 8: 1117–1130 (1994).
Maeda T, Wurglermurphy SM, Saito H: A two-component system that regulates an osmosensing map kinase cascade in yeast. Nature 369: 242–245 (1994).
Mathews S, Lavin M, Sharrock RA: Evolution of the phytochrome gene family and its utility for phylogenetic analyses of angiosperms. Ann Mo Bot Gard 82: 296–321 (1995).
McRee DE, Tainer JA, Meyer TE, Vanbeeumen J, Cusanovich MA, Getzoff ED: Crystallographic structure of a photoreceptor protein at 2.4 Å resolution. Proc Natl Acad Sci USA 86: 6533–6537 (1989).
Morand LZ, Kidd DG, Lagarias JC: Phytochrome levels in the green alga Mesotaenium caldariorum are light regulated. Plant Physiol 101: 97–103 (1993).
Nagasawa S, Tokishita S, Aiba H, Mizuno T: A novel sensor-regulator protein that belongs to the homologous family of signal-transduction proteins involved in adaptive responses in Escherichia coli. Mol Microbiol 6: 799–807 (1992).
Nambu JR, Lewis JO, Wharton KA, Crews ST: The Drosophila single-minded gene encodes a helix-loop-helix protein that acts as a master regulator of CNS midline development Cell 67: 1157–1167 (1991).
Ota IM, Varshavsky A: A yeast protein similar to bacterial 2-component regulators. Science 262: 566–569 (1993).
Pearson WR, Lipman DJ: Improved tools for biological sequence comparison. Proc Natl Acad Sci USA 85: 2444–2448 (1988).
Penfold RJ, Pemberton JM: Sequencing, chromosomal inactivation, and functional expression in Escherichia coli of Ppsr, a gene which represses carotenoid and bacterio-chlorophyll synthesis in Rhodobacter sphaeroides. J Bact 176: 2869–2876 (1994).
Pichersky E, Tanksley SD, Piechulla B, Stayton MM, Dunsmuir P: Nucleotide sequence and chromosomal location of cab-7, the tomato gene encoding the Type II chlorophyll a/b-binding polypeptide of photosystem I. Plant Mol Biol 11: 69–71 (1988).
Provasoli L, Pintner IJ: Artificial media for freshwater algae: problems and suggestions. In: Tryon CA, Hartman RT (eds) The Ecology of Algae, pp. 84–96. University of Pittsburgh, Pittsburgh (1959).
Quail PH: Phytochrome genes and their expression. In: Kendrick RE, Kronenberg GHM (eds) Photomorphogenesis in Plants, 2nd ed., pp. 71–104. Kluwer Academic Publishers, Dordrecht (1994).
Quail PH, Boylan MT, Parks BM, Short TW, Xu Y, Wagner D: Phytochromes: photosensory perception and signal transduction. Science 268: 675–680 (1995).
Reed JW, Chory J: Mutational analyses of light-controlled seedling development in Arabidopsis. Sem Cell Biol 5: 327–334 (1994).
Reed JW, Nagatani A, Elich TD, Fagan M, Chory J: Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol 104: 1139–1149 (1994).
Ropelewski AJ, Nicholas HB: MAXSEGS Optimal Sequence Alignment Program, Pittsburgh (1992).
Ropelewski AJ, Nicholas HB, Gribskov M: PSC-PROFILE-SS V1.0, Pittsburgh (1992).
Sager R, Granick S: Nutritional studies with Chlamydomonas reinhardtii. Ann New York Acad Sci 56: 831–838 (1953).
Sambrook J, Fritsch EF, Maniatis T: Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY (1989).
Schneider-Poetsch HAW: Signal transduction by phytochrome: phytochromes have a module related to the transmitter modules of bacterial sensor proteins. Photochem Photobiol 56: 839–846 (1992).
Schneider-Poetsch HAW, Braun B, Marx S, Schaumburg A: Phytochromes and bacterial sensor proteins are related by structural and functional homologies: hypothesis on phytochrome-mediated signal-transduction. FEBS Lett 281: 245–249 (1991).
Senn G: Die Gestals- und lageveranderung der Pflanzen-Chromatophoren. Verlag von Wilhelm Engelmann, Leipzig (1908).
Sharrock RA, Quail PH: Novel phytochrome sequences in Arabidopsis thaliana: structure, evolution, and differential expression of a plant regulatory photoreceptor family. Genes Devel 3: 1745–1757 (1989).
Shinomura T, Nagatani A, Chory J, Furuya M: The induction of seed germination in Arabidopsis thaliana is regulated principally by phytochrome B and secondarily by phytochrome A. Plant Physiol 104: 363–371 (1994).
Smith H: Phytochrome transgenics: functional, ecological and biotechnological applications. Sem Cell Biol 5: 315–325 (1994).
Smith H, Whitelam GC: Phytochrome, a family of photoreceptors with multiple physiological roles. Plant Cell Environ 13: 695–707 (1990).
Songyang Z, Shoelson SE, Chaudhuri M, Gish G, Pawson T, Haser WG, King F, Roberts T, Rafnofsky S, Lechleider RJ, Neel BG, Birge RB, Fajardo JE, Chou MM, Hanafusa H, Schaffhausen B, Cantley LC: SH2 domains recognize specific phosphopeptide sequences. Cell 72: 767–778 (1993).
Starr RC, Zeikus JA: UTEX—the culture collection of algae at the University of Texas at Austin 1993 List of Cultures. J Phycol 29: 1–106 (1993).
Swofford DL: PAUP—Phylogenetic Analysis Using Parsimony. Illinois Natural History Survey, Champaign (1993).
Taylor AO, Bonner BA: Isolation of phytochrome from the alga Mesotaenium and liverwort Spaerocarpus. Plant Physiol 42: 762–766 (1967).
Thummler F, Dufner M, Kreisl P, Dittrich P: Molecular cloning of a novel phytochrome gene of the moss Ceratodon purpureus which encodes a putative light-regulated protein kinase. Plant Mol Biol 20: 1003–1017 (1992).
Wada M, Grolig F, Haupt W: Light-oriented chloroplast positioning: contribution to progress in photobiology. J Photochem Photobiol B 17: 3–25 (1993).
Wada M, Kadota A: Photomorphogenesis in lower green plants. Annu Rev Plant Physiol Plant Mol Biol 40: 169–191 (1989).
Winands A, Wagner G, Marx S, Schneider-Poetsch HAW: Partial nucleotide sequence of phytochrome from the zygnematophycean green alga Mougeotia. Photochem Photobiol 56: 765–770 (1992).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Lagarias, D.M., Wu, SH. & Lagarias, J.C. Atypical phytochrome gene structure in the green alga Mesotaenium caldariorum . Plant Mol Biol 29, 1127–1142 (1995). https://doi.org/10.1007/BF00020457
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00020457