Abstract
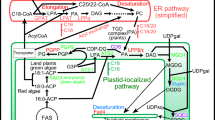
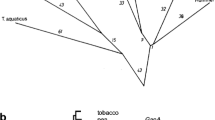
Previous studies indicated that plant nuclear genes for chloroplast and cytosolic isoenzymes of 3-phosphoglycerate kinase (PGK) arose through recombination between a preexisting gene of the eukaryotic host nucleus for the cytosolic enzyme and an endosymbiont-derived gene for the chloroplast enzyme. We readdressed the evolution of eukaryotic pgk genes through isolation and characterisation of a pgk gene from the extreme halophilic, photosynthetic archaebacterium Haloarcula vallismortis and analysis of PGK sequences from the three urkingdoms. A very high calculated net negative charge of 63 for PGK from H. vallismortis was found which is suggested to result from selection for enzyme solubility in this extremely halophilic cytosol. We refute the recombination hypothesis proposed for the origin of plant PGK isoenzymes. The data indicate that the ancestral gene from which contemporary homologues for the Calvin cycle/glycolytic isoenzymes in higher plants derive was acquired by the nucleus from (endosymbiotic) eubacteria. Gene duplication subsequent to separation of Chlamydomonas and land plant lineages gave rise to the contemporary genes for chloroplast and cytosolic PGK isoenzymes in higher plants, and resulted in replacement of the preexisting gene for PGK of the eukaryotic cytosol. Evidence suggesting a eubacterial origin of plant genes for PGK via endosymbiotic gene replacement indicates that plant nuclear genomes are more highly chimaeric, i.e. contain more genes of eubacterial origin, than is generally assumed.
Similar content being viewed by others
Abbreviations
- PGK:
-
3-phosphoglycerate kinase
- FBA:
-
fructose-1,6-bisphosphate aldolase
- GAPDH:
-
glyceraldehyde-3-phosphate dehydrogenase
- TPI:
-
triosephosphate isomerase
References
Alefounder PR, Perham SA: Identification, molecular cloning and sequence analysis of a gene cluster encoding the class II fructose 1,6-bisphosphate aldolase, 3-phosphoglycerate kinase and a putative second glyceraldehyde 3-phosphate dehydrogenase of Escherichia coli. Mol Microbiol 3: 723–732 (1989).
Altekar W, Rajagopalan R: Ribulose bisphosphate carboxylase activity in halophilic archaebacteria. Arch Microbiol 153: 169–174 (1990).
Arcari P, Dello-Russo A, Ianniciello G, Gallo M, Bocchini V: Nucleotide sequence and molecular evolution of the gene coding for glyceraldehyde-3-phosphate dehydrogenase in the thermoacidophilic archaebacterium Sulfolobus solfataricus. Biochem Genet 31: 241–251 (1993).
Benachenhou N, Baldacci G: The gene for a halophilic glutamate dehydrogenase: sequence, transcription analysis and phylogenetic implications. Mol Gen Genet 230: 345–352 (1991).
Brown JR, Doolittle WF: Root of the universal tree of life based on ancient aminoacyl-tRNA synthase gene duplications. Proc Natl Acad Sci USA 92: 2441–2445 (1995).
Cendrin F, Chroboczek J, Zaccia G, Eisenberg H, Mevarech M: Cloning, sequencing and expression in Escherichia coli of the gene coding for malate dehydrogenase from the extremely halophilic archaebacterium Haloarcula marismortui. Biochemistry 23: 4308–4313 (1993).
Chen J-H, Gibson JL, McCue LA, Tabita FR: Identification, expression, and deduced primary structure of transketolase and other enzymes encoded within the form II carbon dioxide fixation operon of Rhodobacter sphaeroides. J Biol Chem 266: 20447–20452 (1991).
Danson MJ: Archaebacteria: the comparative enzymology of their central matabolic pathways. Adv Microbial Physiol 29: 164–231 (1988).
Danson MJ: Central metabolism of the archaebacteria: an overview. Can J Microbiol 35: 58–64 (1989).
Devereux J, Haeberli P, Smithies O: A comprehensive set of sequence analysis programs for the vax. Nucl Acids Res 12: 387–395 (1984).
Dhar NM, Altekar W: Distribution of class I and class II fructose bisphosphate aldolases in halophilic archaebacteria. FEMS Microbiol Lett 35: 177–181 (1986).
Doolittle RF: Of Archae and Eo: What's in a name? Proc Natl Acad Sci USA 92: 2421–2423 (1995).
Fabry S, Heppner P, Dietmaier W, Hensel R: Cloning and sequencing the gene encoding 3-phosphoglycerate kinase from mesophilic Methanobacterium bryantii and thermophilic Methanothermus fervidus. Gene 91: 19–25 (1990).
Fothergill-Gilmore LA, Michels PAM: Evolution of glycolysis. Progr Biophys Mol Biol 59: 105–238 (1993).
Felsenstein J: PHYLIP: Phylogeny Inference Package (version 3.2). Cladistics 5: 164–166 (1989).
Gogarten JP, Kibak H, Dittrich P, Taiz L, Bowman EJ, Bowman BJ, Manolson MF, Poole RJ, Date T, Oshima T, Konishi J, Denda K, Yoshida M: Evolution of the vacuolar H+-ATPase: Implications for the origin of eukaryotes. Proc Natl Acad Sci USA 86: 6661–6665 (1989).
Gupta RS, Singh B: Cloning of the HSP70 gene from Halobacterium marismortui: relatedness of archaebacterial HSP70 to is eubacterial homologs and a model for the evolution of the HSP70 gene. J Bact 174: 4594–4605 (1992).
Hannaert V, Michels P: Structure, function and biogenesis genesis of glycosomes in Kinetoplastida. J Bioenerg Biomembr 20: 205–212 (1994).
Hensel R, Zwickl P, Fabry S, Palm P: Sequence comparison of glyceraldehyde-3-phosphate dehydrogenase from the three urkingdoms: evolutionary implication. Can J Microbiol 35: 81–85 (1989).
Henze K, Badr A, Wettern M, Cerff R, Martin W: A nuclear gene of eubacterial origin in Euglena gracilis reflects cryptic endosymbioses during protist evolution. Proc Natl Acad Sci USA 92: 9122–9126 (1995).
Henze K, Schnarrenberger C, Kellermann J, Martin W: Chloroplast and cytosolic triosephosphate isomerase from spinach: Purification, microsequencing and cDNA sequence of the chloroplast enzyme. Plant Mol Biol 26: 1961–1973 (1994).
Higgins DG, Sharp PM: CLUSTAL: A package for performing multiple sequence alignment on a microcomputer. Gene 73: 237–244 (1988).
Iwabe N, Kuma K-I, Hasegawa M, Osawa S, Miyata T: Evolutionary relationship of archaebacteria, eubacteria and eukaryotes inferred from phylogenetic trees of duplicated genes. Proc Natl Acad Sci USA 86: 9355–9359 (1989).
Jin L, Nei M: Limitations of the evolutionary parsimony method of phylogenetic analysis. Mol Biol Evol 7: 82–102 (1990).
Joshi PB, Dennis PP: Characterization of paralogous and orthologous members of the superoxide dismutase gene family from genera of the halophilic archaebacteria. J Bact 175: 1561–1571 (1993).
Krömer WJ, Arndt E: Halobacterial S9 operon: three ribosomal protein genes are cotranscribed with genes encoding a tRNA-Leu, the enolase, and a putative membrane protein in the archaebacterium Haloarcula (Halobacterium) marismortui. J Biol Chem 266: 24573–24579 (1991).
Kumada Y, Benson DR, Hillemann D, Hosted TJ, Rochefort DA, Thompson CJ, Wohlleben W, Tateno Y: Evolution of the glutamin synthase gene, one of the oldest existing and functioning genes. Proc Natl Acad Sci USA 90: 3009–3013 (1993).
Lam WL, Cohen A, Tsouluhas D, Doolittle WF: Genes for tryptophan biosynthesis in the halophile archaebacterium Haloferax volcanii: the trpDFEG cluster. Proc Natl Acad Sci USA 87: 6614–6618 (1990).
Lam WL, Logan SM, Doolittle WF: Genes for tryptophan biosynthesis in the halophile archaebacterium Haloferax volcanii: the trpDFEG cluster. J Bact 174: 1694–1697 (1992).
Longstaff M, Raines CA, McMorrow EM, Bradbeer JW, Dyer T: Wheat phosphoglycerate kinase: Evidence for recombination between the genes for the chloroplastic and cytosolic enzymes. Nucl Acids Res 17: 6569–6580 (1989).
Ludwig W, Kirchhof G, Klugbauer N, Weizenegger N, Betzl D, Ehrmann M, Hertel C, Jilg S, Tatzel R, Zitelsberger H, Liebl S, Hochberger M, Shah J, Lane D, Wallnöfer P, Scheifer KH: Complete 23S ribosomal RNA sequences of Gram positive bacteria with a low G+C content. System Appl Microbiol 15: 487–501 (1992).
Martin W, Brinkmann H, Savona C, Cerff R: Evidence for a chimaeric nature of nuclear genomes: eubacterial origin of eukaryotic glyceraldehyde-3-phosphate dehydrogenase genes. Proc Natl Acad Sci USA 90: 8692–8696 (1993).
Morse D, Salois P, Markovic P Hastings JW: A nuclear-encoded form II RuBisCO in dinoflagellates. Science 268: 1622–1624 (1995).
Osinga KA, Swinkels BW, Gibson WC, Borst P, Veeneman GH, van Boom JH, Michels PAM, Opperdoes F: Topogenesis of microbody enzymes: a sequence comparison of the genes for the glycosomal (microbody) and cytosolic phosphoglycerate kinases of Trypanosoma brucei. EMBO J, 4: 3811–3817 (1985).
Parker HL, Hill T, Alexander K, Murphy NB, Fish WR, Parsons M: Three genes and two isoenzymes: Gene conversion and the compartmentalization and expression of the phosphoglycerate kinases of Trypanosoma (Nannomonas) congolense. Mol Biochem Parasitol 69: 269–279 (1995).
Pelzer-Reith B, Penger A, Schnarrenberger C: Plant aldolase: cDNA and deduced amino-acid sequence of the chloroplast and cytosol enzyme from spinach. Plant Mol Biol 21: 331–340 (1993).
Prüss B, Meyer HE, Holldorf AW: Characterization of the glyceraldehyde-3-phosphate dehydrogenase from the extremely halophilic archaebacterium Haloarcula vallismortis. Arch Microbiol 160: 5–11 (1993).
Rawal H, Kelkar SM, Altekar W: Ribulose-1,5-bisphosphate dependent CO2 fixation in the halophilic archaebacterium Halobacterium mediterranel. Biochem Biophys Res Comm 156: 451–456 (1988).
Saitou N, Nei M: The neighbor-joining method: a new method for the reconstruction of phylogenetic trees. Mol Biol Evol 4: 406–425 (1987).
Sambrook J, Fritsch EF, Maniatis T: Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York (1989).
Schmidt M, Dvendsen I, Feierabend J: Analysis of the primary structure of the chloroplast isozyme of triose-phosphate isomerase from rye leaves by protein and cDNA sequencing indicates a eukaryotic origin of its gene. Biochim Biophys Acta 1261: 257–264 (1995).
Schnarrenberger C, Flechner A, Martin W: Enzymatic evidence indicating a complete oxidative pentose phosphate in the chloroplasts and an incomplete pathway in the cytosol of spinach leaves. Plant Physiol 108: 609–614 (1995).
Schnarrenberger C, Jacobshagen S, Müller B, Krüger I: Evolution of isozymes of sugar phosphate metabolism in green algae. In: Ogita S, Scandalios JS (eds) Isozymes: Structure, Function, and Use in Biology and Medicine, pp. 743–7460. Wiley-Liss, New York (1990).
Schurig H, Beaucamp N, Ostendorp R, Jaenicke R, Adler E, Knowles JR: Phosphoglycerate kinase and triosephosphate isomerase from the hyperthermophilic bacterium Thermotoga maritima form a covalent bifunctional enzyme complex. EMBO J 14: 442–451 (1995).
Stackebrandt E, Murray RGE, Trüper HG: Proteobacteria classis nov., a name for the phylogenetic taxon that includes the ‘purple bacteria and their relatives’. Int J Syst Bact 38: 321–325 (1988).
Swinkels BW, Evers R, Borst P: The topogenic signal of the glycosomal (microbody) phosphoglycerate kinase of Crithida fasiculata resides in a carboxy-terminal extension. EMBO J 7: 1159–1165 (1988).
Tabita RF: Carbon dioxide fixation and its regulation in cyanobacteria. In: Fay P, van Baalen C (eds) The Cyanobacteria, pp. 95–118 Elsevier, Amsterdam (1987).
Vohra GB, Golding GB, Tsao N, Pearlman RE: A phylogenetic analysis based on the gene encoding phophoglycerate kinase. J Mol Evol 34: 383–395 (1992).
Weeden NF: Genetic and biochemical implications of the endosymbiotic origin of the chloroplast. J Mol Evol 17: 133–139 (1981).
Woese CR: Bacterial evolution. Microbiol Rev 51: 221–271 (1987).
Zillig W, Klenk H-P, Palm P, Leffers H, Pühler G, Gropp F, Garrett RA: Did eukaryotes originate by a fusion event? Endocyt C Res 6: 1–25 (1989).
Zillig W: Comparative biochemistry of Archaea and Bacteria. Curr Opin Genet Devel 1: 544–550 (1991).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Brinkmann, H., Martin, W. Higher-plant chloroplast and cytosolic 3-phosphoglycerate kinases: a case of endosymbiotic gene replacement. Plant Mol Biol 30, 65–75 (1996). https://doi.org/10.1007/BF00017803
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00017803