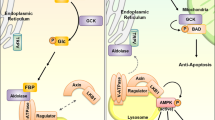
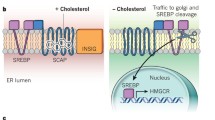
Abstract
Apart from the genetic makeup that plays a fundamental role in health and lifespan determination of an organism, the environment largely influences aging and longevity. One of the environmental factors that determine the well-being of the animal and is involved in various degenerative processes associated with aging is nutrition. Nutritional status in the cell is detected by the interplay of a network of special molecules called nutrient-sensing molecules. These are factors that detect the changes in the nutrient availability of the system and, therefore, integrate various signaling pathways that alter the adaptability of the organism to the changing paradigm to maintain homeostasis. In this present chapter, we will discuss the various mechanisms of nutrient sensing and their interplay in controlling aging and longevity.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Chantranupong L, Wolfson RL, Sabatini DM. Nutrient-sensing mechanisms across evolution. Cell. 2015;161:67–83.
Efeyan A, Comb WC, Sabatini DM. Nutrient sensing mechanisms and pathways. Nature. 2015;517:302–10.
López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–217.
Gallinetti J, Harputlugil E, Mitchell JR. Amino acid sensing in dietary-restriction-mediated longevity: roles of signal-transducing kinases GCN2 and TOR. Biochem J. 2013;449:1–10.
Dong J, Qiu H, Garcia-Barrio M, Anderson J, Hinnebusch AG. Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA-binding domain. Mol Cell. 2000;6:269–79.
Ibba M, Soll D. Aminoacyl-tRNA synthesis. Annu Rev Biochem. 2000;69:617–50.
Diallinas G, Thireos G. Genetic and biochemical evidence for yeast GCN2 protein kinase polymerization. Gene. 1994;143:21–7.
Berlanga JJ, Santoyo J, De Haro C. Characterization of a mammalian homolog of the GCN2 eukaryotic initiation factor 2alpha kinase. Eur J Biochem. 1999;265:754–62.
Scheuner D, Song B, McEwen E, Liu C, Laybutt R, Gillespie P, Saunders T, Bonner-Weir S, Kaufman RJ. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol Cell. 2001;7:1165–76.
Narasimhan J, Staschke KA, Wek RC. Dimerization is required for activation of eIF2 kinase Gcn2 in response to diverse environmental stress conditions. J Biol Chem. 2004;279:22820–32.
Hinnebusch AG. Evidence for translational regulation of the activator of general amino acid control in yeast. Proc Natl Acad Sci USA. 1984;81:6442–6.
Bushman JL, Asuru AI, Matts RL, Hinnebusch AG. Evidence that GCD6 and GCD7, translational regulators of GCN4, are subunits of the guanine nucleotide exchange factor for eIF-2 in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:1920–32.
Wek SA, Zhu S, Wek RC. The histidyl-tRNA synthetase-related sequence in the eIF-2a protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acid. Mol Cell Biol. 1995;15:4497–506.
Munn DH, Sharma MD, Baban B, Harding HP, Zhang Y, Ron D, Mellor AL. GCN2 kinase in T cells mediates proliferative arrest and anergy induction in response to indoleamine 2,3- dioxygenase. Immunity. 2005;22:633–42.
Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485:109–13.
Vlanti A, Rousakis A, Syntichaki P. GCN2 and TOR converge on aging. Aging. 2013;5:584–5.
Yang X, Matsuda K, Bialek P, Jacquot S, Masuoka HC, Schinke T, Li L, Brancorsini S, Sassone-Corsi P, Townes TM, Hanauer A, Karsenty G. ATF4 is a substrate of RSK2 and an essential regulator of osteoblast biology; implication for Coffin-Lowry Syndrome. Cell. 2004;117:387–98.
Deng J, Harding HP, Raught B, Gingras AC, Berlanga JJ, Scheuner D, Kaufman RJ, Ron D, Sonenberg N. Activation of GCN2 in UV-irradiated cells inhibits translation. Curr Biol. 2002;12:1279–86.
Phung TL, Ziv K, Dabydeen D, Eyiah-Mensah G, Riveros M, Perruzzi C, Sun J, Monahan-Earley RA, Shiojima I, Nagy JA, Lin MI, Walsh K, Dvorak AM, Briscoe DM, Neeman M, Sessa WC, Dvorak HF, Benjamin LE. Pathological angiogenesis is induced by sustained Akt signaling and inhibited by rapamycin. Cancer Cell. 2006;10:159–70.
Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, Markhard AL, Sabatini DM. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell. 2006;22:159–68.
Garami A, Zwartkruis FJ, Nobukuni T, Joaquin M, Roccio M, Stocker H, Kozma SC, Hafen E, Bos JL, Thomas G. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol Cell. 2003;11:1457–66.
Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–93.
Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–501.
Hara K, Yonezawa K, Weng QP, Kozlowski MT, Belham C, Avruch J. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J Biol Chem. 1998;273:14484–94.
Avruch J, Long X, Ortiz-Vega S, Rapley J, Papageorgiou A, Dai N. Amino acid regulation of TOR complex 1. Am J Physiol Endocrinol Metab. 2009;296:592–602.
Duran RV, Oppliger W, Robitaille AM, Heiserich L, Skendaj R, Gottlieb E, Hall MN. Glutaminolysis activates Rag-mTORC1 signaling. Mol Cell. 2012;47:349–58.
Kim J, Guan KL. Amino acid signaling in TOR activation. Annu Rev Biochem. 2011;80:1001–32.
Jewell JL, Guan KL. Nutrient signaling to mTOR and cell growth. Trends Biochem Sci. 2013;38:233–42.
Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A, Shi EY, Stumpf CR, Christensen C, Bonham MJ, Wang S, Ren P, Martin M, Jessen K, Feldman ME, Weissman JS, Shokat KM, Rommel C, Ruggero D. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012;485:55–61.
Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature. 2007;450:736–40.
Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10:935–45.
Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303.
Saucedo LJ, Gao X, Chiarelli DA, Li L, Pan D, Edgar BA. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat Cell Biol. 2003;5:566–71.
Long X, Ortiz-Vega S, Lin Y, Avruch J. Rheb binding to mammalian target of rapamycin (mTOR) is regulated by amino acid sufficiency. J Biol Chem. 2005;280:23433–6.
Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM. Ragulator is a GEF for the Rag GTPases that signal amino acid levels to mTORC1. Cell. 2012;150:1196–208.
Bonfils G, Jaquenoud M, Bontron S, Ostrowicz C, Ungermann C, De Virgilio C. Leucyl-tRNA synthetase controls TORC1 via the EGO complex. Mol Cell. 2012;46:105–10.
Han JM, Jeong SJ, Park MC, Kim G, Kwon NH, Kim HK, Ha SH, Ryu SH, Kim S. Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell. 2012;149:410–24.
Duran A, Amanchy R, Linares JF, Joshi J, Abu-Baker S, Porollo A, Hansen M, Moscat J, Diaz-Meco MT. p62 is a key regulator of nutrient sensing in the mTORC1 pathway. Mol Cell. 2011;44:134–46.
Kim YM, Stone M, Hwang TH, Kim YG, Dunlevy JR, Griffin TJ, Kim DH. SH3BP4 is a negative regulator of amino acid-Rag GTPase-mTORC1 signaling. Mol Cell. 2012;46:833–46.
Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 2011;334:678–83.
Kitamoto K, Yoshizawa K, Ohsumi Y, Anraku Y. Dynamic aspects of vacuolar and cytosolic amino acid pools of Saccharomyces cerevisiae. J Bacteriol. 1988;170:2683–6.
Binda M, Péli-Gulli MP, Bonfils G, Panchaud N, Urban J, Sturgill TW, Loewith R, De Virgilio C. The Vam6 GEF controls TORC1 by activating the EGO complex. Mol Cell. 2009;35:563–73.
Neuhaus EM, Almers W, Soldati T. Morphology and dynamics of the endocytic pathway in Dictyostelium discoideum. Mol Biol Cell. 2002;13:1390–407.
Yu L, McPhee CK, Zheng L, Mardones GA, Rong Y, Peng J, Mi N, Zhao Y, Liu Z, Wan F, Hailey DW, Oorschot V, Klumperman J, Baehrecke EH, Lenardo MJ. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature. 2010;465:942–6.
Itoh Y, Kawamata Y, Harada M, Kobayashi M, Fujii R, Fukusumi S, Ogi K, Hosoya M, Tanaka Y, Uejima H, Tanaka H, Maruyama M, Satoh R, Okubo S, Kizawa H, Komatsu H, Matsumura F, Noguchi Y, Shinohara T, Hinuma S, Fujisawa Y, Fujino M. Free fatty acids regulate insulin secretion from pancreatic beta cells through GPR40. Nature. 2003;422:173–6.
Hirasawa A, Tsumaya K, Awaji T, Katsuma S, Adachi T, Yamada M, Sugimoto Y, Miyazaki S, Tsujimoto G. Free fatty acids regulate gut incretin glucagon-like peptide-1 secretion through GPR120. Nat Med. 2005;11:90–49.
Tazoe H, Otomo Y, Kaji I, Tanaka R, Karaki SI, Kuwahara A. Roles of short-chain fatty acids receptors, GPR41 and GPR43 on colonic functions. J Physiol Pharmacol. 2008;2:251–62.
Wang J, Wu X, Simonavicius N, Tian H, Ling L. Medium-chain fatty acids as ligands for orphan G protein-coupled receptor GPR84. J Biol Chem. 2006;281:34457–64.
Laugerette F, Passilly-Degrace P, Patris B, Niot I, Febbraio M, Montmayeur JP, Besnard P. CD36 involvement in orosensory detection of dietary lipids, spontaneous fat preference, and digestive secretions. J Clin Invest. 2005;115:3177–84.
Oh DY, Talukdar S, Bae EJ, Imamura T, Morinaga H, Fan W, Li P, Lu WJ, Watkins SM, Olefsky JM. GPR120 is an omega-3 fatty acid receptor mediating potent anti-inflammatory and insulin-sensitizing effects. Cell. 2010;142:687–98.
Ichimura A, Hirasawa A, Poulain-Godefroy O, Bonnefond A, Hara T, Yengo L, Kimura I, Leloire A, Liu N, Iida K, Choquet H, Besnard P, Lecoeur C, Vivequin S, Ayukawa K, Takeuchi M, Ozawa K, Tauber M, Maffeis C, Morandi A, Buzzetti R, Elliott P, Pouta A, Jarvelin MR, Körner A, Kiess W, Pigeyre M, Caiazzo R, Van Hul W, Van Gaal L, Horber F, Balkau B, Lévy-Marchal C, Rouskas K, Kouvatsi A, Hebebrand J, Hinney A, Scherag A, Pattou F, Meyre D, Koshimizu TA, Wolowczuk I, Tsujimoto G, Froguel P. Dysfunction of lipid sensor GPR120 leads to obesity in both mouse and human. Nat Med. 2012;483:350–4.
Brown AJ, Sun L, Feramisco JD, Brown MS, Goldstein JL. Cholesterol addition to ER membranes alters conformation of SCAP, the SREBP escort protein that regulates cholesterol metabolism. Mol Cell. 2002;10:237–45.
Radhakrishnan A, Sun L-P, Kwon HJ, Brown MS, Goldstein JL. Direct binding of cholesterol to the purified membrane region of SCAP: mechanism for a sterol-sensing domain. Mol Cell. 2004;15:259–68.
Yang T, Espenshade PJ, Wright ME, Yabe D, Gong Y, Aebersold R, Goldstein JL, Brown MS. Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell. 2002;110:489–500.
Song BL, Sever N, DeBose-Boyd RA. Gp78, a membrane-anchored ubiquitin ligase, associates with Insig-1 and couples sterol-regulated ubiquitination to degradation of HMG CoA reductase. Mol Cell. 2005;19:829–40.
Ye J, DeBose-Boyd RA. Regulation of cholesterol and fatty acid synthesis. Cold Spring Harb Perspect Biol. 2011;3:pii: a004754.
Mueckler M, Caruso C, Baldwin SA, Panico M, Blench I, Morris HR, Allard WJ, Lienhard GE, Lodish HF. Sequence and structure of a human glucose transporter. Science. 1985;229:941–5.
Mueckler M. Facilitative glucose transporters. Eur J Biochem. 1994;219:713–25.
Walmsley AR, Barrett MP, Bringaud F, Gould GW. Sugar transporters from bacteria, parasites and mammals: structure-activity relationships. Trends Biochem Sci. 1998;23:476–81.
Thorens B, Mueckler M. Glucose transporters in the 21st century. Am J Physiol Endocrinol Metab. 2010;298:141–5.
Furtado LM, Somwar R, Sweeney G, Niu W, Klip A. Activation of the glucose transporter GLUT4 by insulin. Biochem Cell Biol. 2002;80:569–78.
Watson RT, Kanzaki M, Pessin JE. Regulated membrane trafficking of the insulin-responsive glucose transporter 4 in adipocytes. Endocr Rev. 2004;25:177–204.
Iynedjian PB, Pilot PR, Nouspikel T, Milburn JL, Quaade C, Hughes S, Ucla C, Newgard CB. Differential expression and regulation of the glucokinase gene in liver and islets of Langerhans. Proc Natl Acad Sci USA. 1989;86:7838–42.
Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol. 2012;13:251–62.
Ritter S, Dinh TT, Zhang Y. Localization of hindbrain glucoreceptive sites controlling food intake and blood glucose. Brain Res. 2000;856:37–47.
Frizzell RT, Jones EM, Davis SN, Biggers DW, Myers SR, Connolly CC, Neal DW, Jaspan JB, Cherrington AD. Counterregulation during hypoglycemia is directed by widespread brain regions. Diabetes. 1993;42:1253–61.
Donovan CM. Portal vein glucose sensing. Diabetes Nutr Metab. 2002;15:308–12.
Hevener AL, Bergman RN, Donovan CM. Novel glucosensor for hypoglycemic detection localized to the portal vein. Diabetes. 1997;46:1521–5.
Routh VH. Glucose-sensing neurons: are they physiologically relevant? Physiol Behav. 2002;76:403–13.
McCrimmon RJ, Shaw M, Fan X, Cheng H, Ding Y, Vella MC, Zhou L, McNay EC, Sherwin RS. Key role for AMP-activated protein kinase in the ventromedial hypothalamus in regulating counterregulatory hormone responses to acute hypoglycemia. Diabetes. 2008;57:444–50.
Efeyan A, Zoncu R, Chang S, Gumper I, Snitkin H, Wolfson RL, Kirak O, Sabatini DD, Sabatini DM. Regulation of mTORC1 by the Rag GTPases is necessary for neonatal autophagy and survival. Nature. 2013;493:679–83.
Kahn BB, Alquier T, Carling D, Hardie DG. AMP-activated protein kinase: ancient energy gauge provides clues to modern understanding of metabolism. Cell Metab. 2005;1:15–25.
Cameron KO, Kurumbail RG. Recent progress in the identification of adenosine monophosphate-activated protein kinase (AMPK) activators. Bioorg Med Chem Lett. 2016;26:5139–48.
Chen L, Wang J, Zhang YY, Yan SF, Newmann D, Schattner U, Wang ZX, Wu JW. AMP-activated protein kinase undergoes nucleotide-dependent conformational changes. Nat Struct Mol Biol. 2012;19:716–8.
Hardie DG, Schaffer BE, Brunet A. AMPK: An energy-sensing pathway with multiple inputs and outputs. Trends Cell Biol. 2016;26:190–201.
Sakamoto K, Holman GD. Emerging role for AS160/TBC1D4 and TBC1D1 in the regulation of GLUT4 traffic. Am J Physiol Endocrinol Metab. 2008;295:29–37.
Habets DDJ, Coumans WA, Hasnaoui ME, Zarrinpashneh E, Bertrand L, Viollet B, Kiens B, Jensen TE, Richter EA, Bonen A, Glatz JFC, Luiken JJFP. Crucial role for LKB1 to AMPKα2 axis in the regulation of CD36-mediated long-chain fatty acid uptake into cardiomyocytes. Biochim Biophys Acta. 2009;1791:212–9.
Marsin AS, Bertrand L, Rider MH, Deprez J, Beauloye C, Vincent MF, Van den Berghe G, Carling D, Hue L. Phosphorylation and activation of heart PFK-2 by AMPK has a role in the stimulation of glycolysis during ischaemia. Curr Biol. 2000;10:1247–55.
Merrill GF, Kurth EJ, Hardie DG, Winder WW. AICA riboside increases AMP-activated protein kinase, fatty acid oxidation, and glucose uptake in rat muscle. Am J Phys. 1997;273:1107–12.
Muoio DM, Seefeld K, Witters LA, Coleman RA. AMP-activated kinase reciprocally regulates triacylglycerol synthesis and fatty acid oxidation in liver and muscle: evidence that sn-glycerol-3-phosphate acyltransferase is a novel target. Biochem J. 1999;338:783–91.
Alers S, Löffler AS, Wesselborg S, Stork B. Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy: cross talk, shortcuts, and feedbacks. Mol Cell Biol. 2012;32:2–11.
Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–26.
Puigserver P, Spiegelman BM. Peroxisome proliferator-activated receptor-gamma coactivator 1 alpha (PGC-1 alpha): transcriptional coactivator and metabolic regulator. Endocr Rev. 2003;24:78–90.
Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–8.
Rehman G, Shehzad A, Khan AL, Hamayun M. Role of AMP-activated protein kinase in cancer therapy. Arch Pharm (Weinheim). 2014;347:457–68.
Koch-Nolte F, Fischer S, Haag F, Ziegler M. Compartmentation of NAD+-dependent signalling. FEBS Lett. 2011;585:1651–6.
Fulco M, Schiltz RL, Iezzi S, King MT, Zhao P, Kashiwaya Y, Hoffman E, Veech RL, Sartorelli V. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol Cell. 2003;12:51–62.
Houtkooper RH, Canto C, Wanders RJ, Auwerx J. The secret life of NAD+: an old metabolite controlling new metabolic signaling pathways. Endocr Rev. 2010;31:194–223.
Imai S. Nicotinamide phosphoribosyltransferase (Nampt): a link between NAD biology, metabolism, and diseases. Curr Pharm Des. 2009;15:20–8.
Revollo JR, Grimm AA, Imai S. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J Biol Chem. 2004;279:50754–63.
Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun. 2000;273:793–8.
Haigis MC, Guarente LP. Mammalian sirtuins-emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20:2913–21.
Krejčı A. Metabolic sensors and their interplay with cell signalling and transcription. Biochem Soc Trans. 2012;40:311–23.
Chang HC, Guarente L. SIRT1 and other sirtuins in metabolism. Trends Endocrinol Metab. 2014;25:138–45.
Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol. 2010;5:253–95.
Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, Li Y, Shi J, An W, Hancock SM, He F, Qin L, Chin J, Yang P, Chen X, Lei Q, Xiong Y, Guan KL. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–4.
Guarente L. The logic linking protein acetylation and metabolism. Cell Metab. 2011;14:151–3.
Majaw T, Sharma R. Arginase I expression is upregulated by dietary restriction in the liver of mice as a function of age. Mol Cell Biochem. 2015;407:1–7.
Jakob F, Seefried L, Schwab M. Age and osteoporosis. Effects of aging on osteoporosis, the diagnostics and therapy. Internist (Berl). 2014;55:755–61.
Monfoulet LE, Philippe C, Mercier S, Coxam V, Wittrant Y. Deficiency of G-protein coupled receptor 40, a lipid-activated receptor, heightens in vitro- and in vivo-induced murine osteoarthritis. Exp Biol Med (Maywood). 2015;240:854–66.
Blagosklonny MV, Hall MN. Growth and aging: a common molecular mechanism. Aging. 2009;1:357–62.
Sharma R. Dietary restriction and its multifaceted effects. Curr Sci. 2004;87:1203–10.
Sharma R, Dkhar P. Interventions for healthy aging. In: Sanchetee P, editor. Textbook on geriatric medicine. Hyderabad: Indian Academy of Geriatrics, Paras Medical Publisher; 2014. p. 812–5.
Hadem IKH, Sharma R. Age- and tissue-dependent modulation of IGF-1/PI3K/Akt protein expression by dietary restriction in mice. Horm Metab Res. 2016;48:201–6.
Hadem IKH, Sharma R. Differential regulation of hippocampal IGF-1-associated signaling proteins by dietary restriction in aging mouse. Cell Mol Neurobiol. 2017; https://doi.org/10.1007/s10571-016-0431-7.
Kojima T, Kamei H, Aizu T, Arai Y, Takayama M, Nakazawa S, Ebihara Y, Inagaki H, Masui Y, Gondo Y, Sakaki Y, Hirose N. Association analysis between longevity in the Japanese population and polymorphic variants of genes involved in insulin and insulin-like growth factor 1 signaling pathways. Exp Gerontol. 2004;39:1595–8.
Suh Y, Atzmon G, Cho MO, Hwang D, Liu B, Leahy DJ, Barzilai N, Cohen P. Functionally significant insulin-like growth factor I receptor mutations in centenarians. Proc Natl Acad Sci USA. 2008;105:3438–42.
Anselmi CV, Malovini A, Roncarati R, Novelli V, Villa F, Condorelli G, Bellazzi R, Puca AA. Association of the FOXO3A locus with extreme longevity in a southern Italian centenarian study. Rejuvenation Res. 2009;12:95–104.
Johnson SC, Rabinovitch PS, Kaeberlein M. mTOR is a key modulator of ageing and age-related disease. Nature. 2013;493:338–45.
Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–75.
Hands SL, Proud CG, Wyttenbach A. mTOR’s role in ageing: protein synthesis or autophagy? Aging. 2009;1:586–97.
Ruetenik A, Barrientos A. Dietary restriction, mitochondrial function and aging: from yeast to humans. Biochim Biophys Acta. 2015;1847:1434–47.
Hansen M, Taubert S, Crawford D, Libina N, Lee SJ, Kenyon C. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell. 2007;6:95–110.
Tohyama D, Yamaguchi A, Yamashita T. Inhibition of a eukaryotic initiation factor (eIF2Bdelta/F11A3.2) during adulthood extends lifespan in Caenorhabditis elegans. FASEB J. 2008;22:4327–37.
Dkhar P, Sharma R. Late-onset dietary restriction modulates protein carbonylation and catalase in cerebral hemispheres of aged mice. Cell Mol Neurobiol. 2014;34:307–13.
Mayer C, Grummt I. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene. 2006;25:6384–91.
Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, Kenyon C. A role for autophagy in the extension of lifespan by dietary restriction in C elegans. PLoS Genet. 2008;4:e24.
Tóth ML, Sigmond T, Borsos É, Barna J, Erdélyi P, Takács Vellai K, Orosz L, Kovács AL, Csikós G, Sass M, Vellai T. Longevity pathways converge on autophagy genes to regulate life span in Caenorhabditis elegans. Autophagy. 2008;4:330–8.
Simonsen A, Cumming RC, Brech A, Isakson P, Schubert DR, Finley KD. Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy. 2008;4:176–84.
Alvers AL, Fishwick LK, Wood MS, Hu D, Chung HS, Dunn WAJ, Aris JP. Autophagy and amino acid homeostasis are required for chronological longevity in Saccharomyces cerevisiae. Aging Cell. 2009;8:353–69.
Scott RC, Schuldiner O, Neufeld TP. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell. 2004;7:167–78.
Morck C, Pilon M. C. elegans feeding defective mutants have shorter body lengths and increased autophagy. BMC Dev Biol. 2006;6:39.
Wohlgemuth SE, Julian D, Akin DE, Fried J, Toscano K, Leeuwenburgh C, Dunn JWA. Autophagy in the heart and liver during normal aging and calorie restriction. Rejuvenation Res. 2007;10:281–92.
Cavallini G, Donati A, Gori Z, Pollera M, Bergamini E. The protection of rat liver autophagic proteolysis from the age related decline co-varies with the duration of anti-ageing food restriction. Exp Gerontol. 2001;36:497–506.
Panowski SH, Wolff S, Aguilaniu H, Durieux J, Dillin A. PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature. 2007;447:550–5.
Wauquier F, Philippe C, Léotoing L, Mercier S, Davicco MJ, Lebecque P, Guicheux J, Pilet P, Miot-Noirault E, Poitout V, Alquier T, Coxam V, Wittrant Y. The free fatty acid receptor G protein-coupled receptor 40 (GPR40) protects from bone loss through inhibition of osteoclast differentiation. J Biol Chem. 2013;288:6542–51.
Martini C, Pallottini V, Cavallini G, Donati A, Bergamini E, Trentalance A. Caloric restrictions affect some factors involved in age-related hypercholesterolemia. J Cell Biochem. 2007;101:235–43.
Marino M, Pallottini V, D’Eramo C, Cavallini G, Bergamini E, Trentalance A. Age-related changes of cholesterol and dolichol biosynthesis in rat liver. Mech Ageing Dev. 2002;123:1183–9.
Pallottini V, Montanari L, Cavallini G, Bergamini E, Gori Z, Trentalance A. Mechanisms underlying the impaired regulation of 3-hydroxy-3-methylglutaryl coenzyme A reductase in aged rat liver. Mech Ageing Dev. 2004;125:633–9.
Smiljanic K, Vanmierlo T, Djordjevic AM, Perovic M, Ivkovic S, Lütjohann D, Kanazir S. Cholesterol metabolism changes under long-term dietary restrictions while the cholesterol homeostasis remains unaffected in the cortex and hippocampus of aging rats. Age (Dordr). 2014;36:9654.
Carayannopoulos MO, Chi MM, Cui Y. GLUT8 is a glucose transporter responsible for insulin-stimulated glucose uptake in the blastocyst. Proc Natl Acad Sci USA. 2000;97:7313–8.
Doege H, Schürmann A, Bahrenberg G, Brauers A, Joost HG. GLUT8, a novel member of the sugar transport facilitator family with glucose transport activity. J Biol Educ. 2000;275:16275–80.
Banerjee A, Anuradha, Mukherjee K, Krishna A. Testicular glucose and its transporter GLUT 8 as a marker of age-dependent variation and its role in steroidogenesis in mice. J Exp Zool A Ecol Genet Physiol. 2014;321:490–502.
Cho SJ, Moon JS, Lee CM, Choi AM, Stout-Delgado HW. GLUT-1-dependent glycolysis is increased during aging-related lung fibrosis and phloretin inhibits lung fibrosis. Am J Respir Cell Mol Biol. 2016; https://doi.org/10.1165/rcmb.2016-0225OC.
Novelli M, De Tata V, Bombara M, Bergamini E, Masiello P. Age-dependent reduction in GLUT-2 levels is correlated with the impairment of the insulin secretory response in isolated islets of Sprague-Dawley rats. Exp Gerontol. 2000;35:641–51.
Torres TP, Catlin RL, Chan R, Fujimoto Y, Sasaki N, Printz RL, Newgard CB, Shiota M. Restoration of hepatic glucokinase expression corrects hepatic glucose flux and normalizes plasma glucose in zucker diabetic fatty rats. Diabetes. 2009;58:78–86.
Kharbhih WJ, Sharma R. Inorganic pyrophosphatase of cardiac and skeletal muscle is enhanced by dietary restriction in mice during aging. In: Rath PC, Sharma R, Prasad S, editors. Topics in biomedical gerontology. Singapore: Springer Nature; 2017. p. 117–22.
Steinberg GR, Kemp BE. AMPK in health and disease. Physiol Rev. 2009;89:1025–78.
Petersen KF, Befroy D, Dufour S, Dziura J, Ariyan C, Rothman DL, DiPietro L, Cline GW, Shulman GI. Mitochondrial dysfunction in the elderly: possible role in insulin resistance. Science. 2003;300:1140–2.
Ma Y, Li J. Metabolic shifts during aging and pathology. Compr Physiol. 2015;5:667–86.
Onken B, Driscoll M. Metformin induces a dietary restriction-like state and oxidative stress response to extend C. elegans healthspan via AMPK, LKB1, and SKN-1. PLoS ONE. 2010; https://doi.org/10.1371/journal.pone.0008758.
Funakoshi M, Tsuda M, Muramatsu K, Hatsuda H, Morishita S, Aigaki T. A gain-of-function screen identifies wdb and lkb1 as lifespan-extending genes in Drosophila. Biochem Biophys Res Commun. 2011;405:667–72.
Viollet B, Andreelli F, Jørgensen SB, Perrin C, Flamez D, Mu J, Wojtaszewski JF, Schuit FC, Birnbaum M, Richter E, Burcelin R, Vaulont S. Physiological role of AMP-activated protein kinase (AMPK): insights from knockout mouse models. Biochem Soc Trans. 2003;31:216–9.
Reznick RM, Zong H, Li J, Morino K, Moore IK, Yu HJ, Liu ZX, Dong J, Mustard KJ, Hawley SA, Befroy D, Pypaert M, Hardie DG, Young LH, Shulman GI. Aging-associated reductions in AMP-activated protein kinase activity and mitochondrial biogenesis. Cell Metab. 2007;5:151–6.
Salminen A, Kaarniranta K. AMP-activated protein kinase (AMPK) controls the aging process via an integrated signaling network. Ageing Res Rev. 2012;11:230–41.
Imai S, Guarente L. It takes two to tango: NAD+ and sirtuins in aging/longevity control. Aging Mech Dis. 2016;2:16017. https://doi.org/10.1038/npjamd.2016.17.
Hadem IKH. Dietary and age-dependent regulation of insulin-like growth factor-1 and its related signaling in mice. PhD dissertation, North-Eastern Hill University, Shillong, India. 2016.
Satoh A, Brace CS, Rensing N, Cliften P, Wozniak DF, Herzog ED, Yamada KA, Imai S. Sirt1 extends life span and delays aging in mice through the regulation of Nk2 homeobox 1 in the DMH and LH. Cell Metab. 2013;18:416–30.
Giblin W, Skinner ME, Lombard DB. Sirtuins: guardians of mammalian healthspan. Trends Genet. 2014;30:271–86.
Kanfi Y, Naiman S, Amir G, Peshti V, Zinman G, Nahum L, Bar-Joseph Z, Cohen HY. The sirtuin SIRT6 regulates lifespan in male mice. Nature. 2012;483:218–21.
Gomes AP, Price NL, Ling AJ, Moslehi JJ, Montgomery MK, Rajman L, White JP, Teodoro JS, Wrann CD, Hubbard BP, Mercken EM, Palmeira CM, De Cabo R, Rolo AP, Turner N, Bell EL, Sinclair DA. Declining NAD(+) induces a pseudohypoxic state disrupting nuclear-mitochondrial communication during aging. Cell. 2013;155:1624–38.
Stein LR, Imai S. Specific ablation of Nampt in adult neural stem cells recapitulates their functional defects during aging. EMBO J. 2014;33:1321–40.
Ghosh D, Levault KR, Brewer GJ. Relative importance of redox buffers GSH and NAD(P)H in age-related neurodegeneration and Alzheimer disease-like mouse neurons. Aging Cell. 2014;13:631–40.
Jukarainen S, Heinonen S, Rämö JT, Rinnankoski-Tuikka R, Rappou E, Tummers M, Muniandy M, Hakkarainen A, Lundbom J, Lundbom N, Kaprio J, Rissanen A, Pirinen E, Pietiläinen KH. Obesity is associated with low NAD/SIRT pathway expression in adipose tissue of BMI-discordant monozygotic twins. J Clin Endocrinol Metab. 2015;101:275–83.
Lin SJ, Ford E, Haigis M, Liszt G, Guarente L. Calorie restriction extends yeast life span by lowering the level of NADH. Genes Dev. 2004;18:12–6.
Bai P, Cantó C, Oudart H, Brunyánszki A, Cen Y, Thomas C, Yamamoto H, Huber A, Kiss B, Houtkooper RH, Schoonjans K, Schreiber V, Sauve AA, Menissier-de Murcia J, Auwerx J. PARP-1 inhibition increases mitochondrial metabolism through SIRT1 activation. Cell Metab. 2011;13:461–8.
Imai S, Guarente L. NAD+ and sirtuins in aging and disease. Trends Cell Biol. 2014;24:464–71.
Mouchiroud L, Houtkooper RH, Moullan N, Katsyuba E, Ryu D, Canto C, Mottis A, Jo YS, Viswanathan M, Schoonjans K, Gaurente L, Auwerx J. The NAD+/Sirtuin pathway modulates longevity through activation of mitochondrial UPR and FOXO signalling. Cell. 2013;154:430–41.
Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng HL, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–5.
Papa L, Germain D. SirT3 regulates the mitochondrial unfolded protein response. Mol Cell Biol. 2014;34:699–710.
Baur JA, Ungvari Z, Minor RK, Le Couteur DG, de Cabo R. Are sirtuins viable targets for improving healthspan and lifespan? Nat Rev Drug Discov. 2012;11:443–61.
Hebert AS, Dittenhafer-Reed KE, Yu W, Bailey DJ, Selen ES, Boersma MD, Carson JJ, Tonelli M, Balloon AJ, Higbee AJ, Westphall MS, Pagliarini DJ, Prolla TA, Assadi-Porter F, Roy S, Denu JM, Coon JJ. Calorie restriction and SIRT3 trigger global reprogramming of the mitochondrial protein acetylome. Mol Cell. 2013;49:186–99.
Sundaresan NR, Gupta M, Kim G, Rajamohan SB, Isbatan A, Gupta MP. Sirt3 blocks the cardiac hypertrophic response by augmenting Foxo3a-dependent antioxidant defense mechanisms in mice. J Clin Invest. 2009;119:2758–71.
Song J, Ke SF, Zhou CC, Zhang SL, Guan YF, Xu TY, Sheng CQ, Wang P, Miao CY. Nicotinamide phosphoribosyltransferase is required for the calorie restriction-mediated improvements in oxidative stress, mitochondrial biogenesis, and metabolic adaptation. J Gerontol A Biol Sci Med Sci. 2014;69:44–57.
Cartee GD, Dean DJ. Glucose transport with brief dietary restriction: heterogenous responses in muscles. Am J Phys. 1994;266:946–52.
Wang H, Wu H, Guo H, Zhang G, Zhang R, Zhan S. Increased hypoxia-inducible factor 1alpha expression in rat brain tissues in response to aging. Neural Regen Res. 2012;7:778–82.
Chen TT, Maevsky EI, Uchitel ML. Maintenance of homeostasis in the aging hypothalamus: the central and peripheral roles of succinate. Front Endocrinol (Lausanne). 2015;6:7.
Sheedfar F, Sung MM, Aparicio-Vergara M, Kloosterhuis NJ, Miquilena-Colina ME, Vargas-Castrillón J, Febbraio M, Jacobs RL, de Bruin A, Vinciguerra M, García-Monzón C, Hofker MH, Dyck JR, Koonen DP. Increased hepatic CD36 expression with age is associated with enhanced susceptibility to nonalcoholic fatty liver disease. Aging (Albany NY). 2014;6:281–95.
Abumrad NA, el Maghrabi MR, Amri EZ, Lopez E, Grimaldi PA. Cloning of a rat adipocyte membrane protein implicated in binding or transport of longchain fatty acids that is induced during preadipocyte differentiation. Homology with human CD36. J Biol Chem. 1993;268:17665–8.
Miyauchi S, Hirasawa A, Iga T, Liu N, Itsubo C, Sadakane K, Hara T, Tsujimoto G. Distribution and regulation of protein expression of the free fatty acid receptor GPR120. Naunyn Schmiedeberg’s Arch Pharmacol. 2009;379:427–34.
Acknowledgments
Authors thank the Department of Biochemistry, North-Eastern Hill University, Shillong, for the research facilities. We thank the UGC-SAP program in Biochemistry, the DBT Infrastructure, and Biotech Hub to NEHU for financial support.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Hadem, I.K.H., Majaw, T., Sharma, R. (2020). Interplay Between Nutrient-Sensing Molecules During Aging and Longevity. In: Rath, P. (eds) Models, Molecules and Mechanisms in Biogerontology. Springer, Singapore. https://doi.org/10.1007/978-981-32-9005-1_18
Download citation
DOI: https://doi.org/10.1007/978-981-32-9005-1_18
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-32-9004-4
Online ISBN: 978-981-32-9005-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)