Abstract
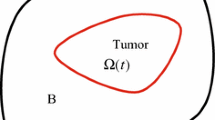
The biochemical dynamics involved in tumor growth can be broadly classified into three distinct spatial scales: the tumor scale, the cell-ECM interactions and the sub-cellular processes. This work presents the tumor scale investigations, which are expected to eventually lead to a system-level understanding of the progression of cancer. Many of the macroscopic phenomena of physiological relevance, such as tumor size and shape, formation of necrotic core and vascularization, proliferation and metastasis of cell populations, external mechanical interactions, etc., can be treated within a continuum framework by modeling the evolution of various species involved by transport equations coupled with mechanics. This framework is an extension of earlier work (Garikipati et al. in J. Mech. Phys. Solids 52:1595–1625, 2004; Narayanan et al. in Biomech. Model. Mechanobiol. 8:167–181, 2009, J. Phys. Condens. Matter. 22:194122, 2010) based on the continuum theory of mixtures for modeling biological growth. Specifically, the focus is on demonstrating the effectiveness of mechano-transport coupling in simulating tumor growth dynamics and explaining some in vitro observations like mechanics-induced ellipsoidal tumor shapes. Additionally, this work also seeks to demonstrate the effectiveness of tools like adaptive mesh refinement and automatic differentiation in handling highly nonlinear, coupled multiphysics systems.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
Notes
- 1.
Automatic differentiation, also referred to as algorithmic differentiation, calculates derivatives of functions up to any order to within machine precision by reducing complex functions to elementary arithmetic operations and elementary functions by repeated application of the chain rule. It can result in significant speedup of multiphysics implementations by computing the Jacobian of finite element residuals. For variational problems, even greater ease of implementation is possible: only the energy functional needs to be coded, and the system of residual equations and the Jacobian can be computed by taking variational derivatives of the functional and residual equations, respectively.
References
Bangerth W, Hartmann R, Kanschat G (2007) deal.II–a general purpose object oriented finite element library. ACM Trans Math Softw 33:24
Bourrat-Floeck B, Groebbe K, Mueller-Klieser W (1991) Biological response of multicellular EMT6 spheroids to exogenous lactate. Int J Cancer 47:792–799
Casciari J, Sotirchos S, Sutherland R (1992) Variations in tumor cell growth rates and metabolism with oxygen concentration, glucose concentration, and extracellular pH. J Cell Physiol 151:386–394
Garikipati K, Arruda EM, Grosh K, Narayanan H, Calve SC (2004) A continuum treatment of growth in biological tissue: the coupling of mass transport and mechanics. J Mech Phys Solids 52:1595–1625
Mills KL, Garikipati K, Kemkemer R (2011) Experimental characterization of tumor spheroids for studies of the energetics of tumor growth. Int J Mater Res 7:889–895
Mills K, Kemkemer R, Rudraraju S, Garikipati K (2012) The growth of human colon adenocarcinoma tumors in agarose hydrogels (in preparation)
Narayanan H, Arruda EM, Grosh K, Garikipati K (2009) The micromechanics of fluid-solid interactions during growth in porous soft biological tissue. Biomech Model Mechanobiol 8:167–181
Narayanan H, Verner S, Mills K, Kemkemer R, Garikipati K (2010) In silico estimates of the free energy rates in growing tumor spheroids. J Phys Condens Matter 22:194122
Sacado (2011) Automatic differentiation tools for C++ applications. The Trilinos Project
Valastyan S, Weinberg RA (2011) Tumor metastasis: molecular insights and evolving paradigms. Cell 147:275–292
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2013 Springer Science+Business Media Dordrecht
About this paper
Cite this paper
Rudraraju, S., Mills, K.L., Kemkemer, R., Garikipati, K. (2013). Multiphysics Modeling of Reactions, Mass Transport and Mechanics of Tumor Growth. In: Holzapfel, G., Kuhl, E. (eds) Computer Models in Biomechanics. Springer, Dordrecht. https://doi.org/10.1007/978-94-007-5464-5_21
Download citation
DOI: https://doi.org/10.1007/978-94-007-5464-5_21
Publisher Name: Springer, Dordrecht
Print ISBN: 978-94-007-5463-8
Online ISBN: 978-94-007-5464-5
eBook Packages: EngineeringEngineering (R0)