Abstract
The symbiosis between Vibrio fischeri and the Hawaiian squid Euprymna scolopes has been intensively studied for over 20 years. Although V. fischeri is a cosmopolitan marine bacterium, the experimental tractability of this symbiosis has engendered the most research interest. The utility of this model system is clear from its demonstrated ability to inform our understanding of the specificity of bacteria-host interactions, the initiation and persistence of such interactions, the influence of bacteria on host development, the evolution of facultatively symbiotic bacteria, and the genes and regulators that underlie successful infection. Investigations of this symbiosis have revealed that the two organisms engage in a dynamic interaction that begins when newly hatched juvenile squid are exposed to V. fischeri cells. Positive and negative factors expressed by the squid—in some cases in response to bacterial products—ensure colonization only by V. fischeri. Colonization by V. fischeri triggers developmental events in the host, many of which minimize subsequent colonization by other bacteria or superinfection by competing V. fischeri strains. Growth of V. fischeri to high cell density occurs within specific internal symbiotic tissues in the light organ, and results in the production by V. fischeri of bioluminescence, the product of the symbiosis. The control of bioluminescence in V. fischeri has become a paradigmatic model of pheromone-mediated regulation, and the discovery of other regulators has likewise elucidated how bacteria switch between free-living and mutualistic lifestyles. Finally, a daily expulsion of the majority of symbiotic bacteria results in a cycle of growth resembling a laboratory “batch” culture, and experimental approaches are being employed to study the physiology of V. fischeri with relevance to its natural niche. A number of bacterial genes and traits necessary for successful colonization have been identified and characterized, with intriguing parallels to pathogenic bacteria. This chapter describes what is currently known about this symbiosis and the roles of bacterial factors in promoting colonization.
An Erratum to this article can be found at http://dx.doi.org/10.1007/978-3-642-30194-0_118
You have full access to this open access chapter, Download reference work entry PDF
Similar content being viewed by others
Introduction
Overview
The marine bioluminescent bacterium Vibrio fischeri forms a highly specific mutualistic symbiosis with the Hawaiian bobtail squid Euprymna scolopes. The study of this symbiosis over the past 20 years has been aided by the nature of the interaction itself: the squid hatch without V. fischeri but rapidly acquire them from the seawater, and thus experimentally, wild-type or mutant bacteria can be added to the seawater and the process of colonization examined. Once the bacteria colonize, they bioluminesce, and this natural light production provides a noninvasive means of monitoring colonization. Furthermore, tools such as green fluorescent protein (GFP) have been engineered to permit visualization of bacteria at all stages of colonization in the transparent symbiotic tissue of juvenile squid. The bacterium can be readily manipulated genetically, and the genome sequences of multiple strains are known, making it feasible to test specific genes for their roles in bacteria-host interactions. Finally, investigations into the biology of the squid and its symbiotic organ, the light organ, provide a framework for developing hypotheses to be tested. The result is a robust model that is continually yielding novel insights.
In this chapter, we describe in detail the biology of V. fischeri as it relates to the ability of this microbe to form a specific symbiosis with E. scolopes. We begin with an introduction to the ecology of V. fischeri and its squid host. Then, to provide a basis for understanding the symbiosis, we describe the structure of the symbiotic light organ and give an overview of what is known about the dynamics of colonization and specificity in the symbiosis. We then discuss host development and the known roles of the bacteria and bacterial signals in developmental processes. With this foundation, we then describe a number of bacterial genes and phenotypes whose roles in symbiosis have been investigated, including the processes of bacterial bioluminescence, biofilm formation, motility, and iron uptake. Finally, we conclude with a brief discussion of evolution and our perspectives on the field and its future.
Ecology of V. fischeri and Its Squid Host E. scolopes
It is not possible to appreciate the biology of V. fischeri fully without first understanding the environments that it experiences during its life cycle. As a marine bacterium, V. fischeri primarily resides in seawater, which contains dissolved salts at a concentration of 3.5%. The dissolved salts include sodium, chloride, magnesium, sulfate, calcium, and potassium. In this environment, V. fischeri can be found free-living in seawater and also associated with sediment (Lee and Ruby 1992, 1994b). It also can be found colonizing animal hosts.
The best known of these animal associations are exquisitely evolved light-organ symbioses in which V. fischeri colonizes the light-emitting organs of certain fishes and squids, generating bioluminescence used by the host in exchange for nutrients and a privileged niche. For example, V. fischeri colonizes light organs in monocentrid “pinecone” fishes of the genera Cleidopus or Monocentris, both found in the Pacific Ocean (Fitzgerald 1977; Ruby and Nealson 1976), and in sepiolid “bobtail” squids of the genera Sepiola or Euprymna, which are found in the Mediterranean Sea and Pacific Ocean, respectively (Fidopiastis et al. 1998; Jones et al. 2006; Nishiguchi 2002; Nishiguchi et al. 1998; Wei and Young 1989). Although its role as a bioluminescent symbiont is well studied, the association of V. fischeri with hosts is not restricted to monospecific light-organ symbioses. It has also been isolated from multi-species gut consortia of fish (Ramesh and Venugopalan 1989; Ruby and Morin 1979; Sugita and Ito 2006) and from chitinous structures on the invertebrate hydrozoans Aglaophenia tubiformis and Halopteris diaphana (Stabili et al. 2008).
Although it is a close associate of marine animals, the genetic and physiological capacity of V. fischeri is unlike that of obligate symbionts (Ochman and Moran 2001; Ruby et al. 2005). Both its demonstrated metabolic flexibility and its genomic content indicate that V. fischeri is able to grow under a range of conditions using any of several substrates, unlike many obligate symbionts that evolve reduced genomes adapted to a relatively simple and constant host environment. The metabolic diversity and genomic content of V. fischeri suggest that an important component of this bacterium’s life history occurs outside of specific symbioses, consistent with the observation that V. fischeri has been found free-living in different marine environments, both aerobic and anaerobic, in the water column and in sediments (Garcia-Amado et al. 2011; Jones et al. 2007; Lee and Ruby 1994b; Orndorff and Colwell 1980; Ruby et al. 1980). While it is possible that V. fischeri isolated from sediments and the water column had recently cycled through a host, it also has been isolated in regions far from any known light-organ symbioses, such as the Sargasso Sea and coastal waters off the northeastern United States.
Despite its frequent association with hosts and its phylogenetic relationship to known pathogens such as V. cholerae, V. parahaemolyticus, and V. vulnificus (Tantillo et al. 2004), to our knowledge, V. fischeri has never been documented as a pathogen. Its inability to grow at 37 °C certainly restricts it from causing human infections, and it has not been observed to cause disease in marine organisms even at permissive temperatures. The apparent nonpathogenicity of V. fischeri contrasts with its relative Vibrio salmonicida, which causes cold-water vibriosis in salmonids, and with another common bioluminescent marine bacterium, Vibrio harveyi, which apparently is responsible for some die-off events in aquacultured shrimp. V. fischeri has been isolated from aquaculture tanks in fish-rearing facilities (Alcaide 2003; Montes et al. 2006), and in one instance, it was isolated from organs of diseased aquacultured fish (Lamas et al. 1990); however, it was not shown to be causal to morbidity or mortality. It seems plausible that in this isolated incident, V. fischeri may have been a secondary opportunist flourishing in a host already compromised by another microbe. Overall, although the V. fischeri genome encodes homologs of virulence factors found in other members of the Vibrionaceae (Ruby et al. 2005), this species appears to enter benign or beneficial associations with hosts.
Among its specialized light-organ symbioses, the best studied is that between V. fischeri and E. scolopes, the Hawaiian bobtail squid (Fig. 20.1a ). E. scolopes is a nocturnal predator that feeds on polychaetes and shrimp. The hatchlings (Fig. 20.2a ) are typically only ~3–4 mm long but can grow to be ten times that length (Moynihan 1983; Shears 1988). The adults likely live less than a year in the wild (Hanlon et al. 1997; Singley 1983) and are found near the Hawaiian coast in shallow, sandy reef areas, in a few meters or even as little as a few centimeters of water. It is unclear the extent to which this habitat reflects where they live versus where researchers typically search for them. There are reports of E. scolopes being found well outside the reefs and even at depths of 200 m (Berry 1912), but for convenience, researchers typically have stayed closer to shore. These animals appear to be solitary except when they are mating, which has been observed both in shallow water and in captivity (Fig. 20.1b ). The ability to maintain and mate E. scolopes in laboratory aquaria has underpinned its development as a model experimental system.
Adult E. scolopes. (a) Adult E. scolopes sitting on coral sand; (b) a mating pair of E. scolopes; (c) sand-covered E. scolopes; (d) ventrally dissected E. scolopes; (e) a close-up view of the adult light organ (Image from panel a was taken from Stabb and Millikan (2009), while images from b and c are courtesy of Kati Geszvain)
Juvenile E. scolopes. (a) Juvenile E. scolopes, with the light organ prominent as a black shape in the center of the mantle; (b) an image, generated with scanning electron microscopy, in which the ciliated epithelial appendages are seen extending from the surface and the underlying light-organ tissues are apparent; (c) a cross section of one of the appendages revealing a blood sinus; (d) the surface of part of the light organ depicting three pores, in/near one of which is an aggregate of V. fischeri cells; (e) cartoon depicting the structure of one-half of the juvenile light organ with three crypts labeled (1, 2, and 3); (f) cross section of a region of a colonized crypt. mv microvilli, ep epithelium (The image from panel a was previously published (Dunn and Stabb 2008a), and the image in panel d is cropped from a version that was previously published (Yip et al. 2006))
After mating, E. scolopes females lay clutches of eggs that lack V. fischeri symbionts. Upon hatching, each new generation must acquire V. fischeri from the surrounding seawater (Wei and Young 1989), a phenomenon that allows researchers in controlled laboratory environments to compare animals infected with different strains or with no V. fischeri symbionts at all. If V. fischeri is present, infection occurs within hours, and it is so efficient that no uninfected E. scolopes has ever been found in the wild. The animal maintains a monospecific culture of V. fischeri in its light organ. As discussed below, this specific infection with V. fischeri triggers a developmental program in the light-organ tissue. Although the light organ undergoes large morphological changes, the animals maintain a monospecific culture of V. fischeri throughout their life, allowing the squid to exploit the symbionts’ bioluminescence.
Whether to hide from predators or prey, E. scolopes makes extensive use of camouflaging (Anderson and Mather 1996; Shears 1988), a general strategy that appears to include their use of V. fischeri bioluminescence. The animals cover themselves with sand and use chromatophores to change colors among a natural-looking palette (Fig. 20.1c ). They can even be observed swimming with a sand coat, which they can discard quickly (Shears 1988). Similarly, the V. fischeri symbionts are apparently used in a strategy referred to as “counterillumination,” where ventrally directed bioluminescence is used to obscure the squid’s silhouette from organisms beneath it in the water column. The strongest evidence that this is the function of symbiotic bioluminescence includes the architecture of the organ (McFall-Ngai and Montgomery 1990) and the observation that the light emitted is controlled to directly correlate with the ambient downwelling light (Jones and Nishiguchi 2004). Although a nutritional or other benefit of the symbionts cannot be ruled out, E. scolopes raised through a complete life cycle without V. fischeri did not appear compromised (Claes and Dunlap 2000), further reinforcing the idea that symbiont bioluminescence is the main advantage to the host. The counterillumination model above and other possible explanations for the use of bioluminescent symbionts by the squid were reviewed recently (Stabb and Millikan 2009).
The advantage of the symbiosis for V. fischeri appears clearer. V. fischeri is provided nutrients in the E. scolopes light organ, and this supports its rapid growth (Ruby and Asato 1993). Furthermore, V. fischeri cells also appear to benefit from the host immune system, which maintains an exclusive relationship with the bacteria, protecting them from predation or competition by other microbes. Each morning, the squid expel most of the V. fischeri cells in their light organ out into the environment and then support regrowth of the remaining symbionts throughout the day (Boettcher et al. 1996) (Fig. 20.3 ). Given this daily venting and re-culturing, one would expect to find relatively high populations of V. fischeri in habitats occupied by E. scolopes, which indeed has been observed (Lee and Ruby 1994b). Other ecological studies (Lee and Ruby 1994a) support the idea that in shallow, sandy Hawaiian reefs occupied by E. scolopes, the ability to colonize this squid is advantageous for V. fischeri. Taken together, the evidence suggests that this symbiosis is a mutualism that benefits both partners.
Given the evidence for mutualism, it is not surprising that V. fischeri isolates from E. scolopes appear to have coevolved with this host. There is considerable evidence that certain strains of V. fischeri have adapted to be especially proficient colonizers of E. scolopes (Lee and Ruby 1994a; Mandel et al. 2009; Nishiguchi 2002; Nishiguchi et al. 1998; Schuster et al. 2010), and repeated passage of strains through E. scolopes in the laboratory has shown that less proficient colonizers can evolve into more effective symbionts (Schuster et al. 2010). Interestingly, the gene encoding the regulatory sensor RscS, discussed further below, appears to be a key genetic acquisition in the evolution of V. fischeri, leading to more proficient colonization of Euprymna hosts in the Pacific (Mandel et al. 2009).
As a coevolved mutualism, the V. fischeri-E. scolopes symbiosis resembles many specific bacterium-host interactions found in nature. Given several features that make it experimentally tractable, it serves as a powerful natural model for such associations. Although V. fischeri may not require E. scolopes or other light-organ symbioses to survive and grow in the environment, in locales where hosts are available, light-organ colonization appears to be very important in its ecology. Thus, studies of V. fischeri infecting E. scolopes address issues directly significant to the ecology of the bacterium in nature, and they can elucidate our understanding of bacterium-host symbioses in general.
Structure of Light Organ, Dynamics of Colonization, and Development
Structure of Light Organ
In adult E. scolopes, V. fischeri cells reside within a complex, bilobed organ at a level in excess of 109 bacteria or approximately 1011 cells per ml of light-organ fluid (Boettcher and Ruby 1990; Nyholm and McFall-Ngai 1998) (Fig. 20.1d ). It is at these high cell densities that the bacterial contribution to the symbiosis, bioluminescence, is produced. The adult organ contains several tissues, including lens and reflector tissues that direct and modulate the light (Fig. 20.1e ). Of note, the light organ occupies a significant portion of the space within the squid’s body cavity (mantle), a feature that suggests the relative importance of this organ and the symbiosis to the life cycle of the animal.
Juvenile E. scolopes, which hatch without symbionts (aposymbiotic), are first exposed to V. fischeri cells when the animal ventilates seawater into its mantle cavity. Derived from an outgrowth of the digestive tract (Montgomery and McFall-Ngai 1993), the juvenile light organ (Fig. 20.2b & e ) features two sets of ciliated surface appendages that project into the mantle cavity (McFall-Ngai and Ruby 1991). The cilia on these appendages, along with the cilia decorating ridges on either side of the organ, entrain the bacteria-containing ambient seawater toward pores that serve as the entrances to the light organ (McFall-Ngai and Ruby 1998) (Fig. 20.2d & e ). In addition to the cilia, the surface of the light organ is coated with mucus that is secreted from the epithelial cells that line the appendages (Nyholm et al. 2000, 2002). The directed movement of the cilia and surface-secreted mucus are thought to promote attachment of V. fischeri carried into the squid with the ventilated seawater. Thus, at a very early stage in colonization, V. fischeri experiences a mucus-coated surface, an environment that is vastly different from seawater.
A small aggregate of V. fischeri cells accumulates on the surface of the symbiotic light organ, then ultimately individual cells track into pores to enter the organ (Fig. 20.2d ) (Nyholm et al. 2000). A total of six pores exist, three on each side of the organ. They range in size from 5 to 15 μm in diameter (Montgomery and McFall-Ngai 1993). Thus, V. fischeri cells, which are approximately 1–2 μm in length (Millikan and Ruby 2003; Ruby and Asato 1993), are substantially smaller than the pores through which they enter. Bacterial motility appears to be important for entry, as nonmotile bacteria appear to aggregate but do not migrate to the pores (Nyholm et al. 2000).
The six pores open into six ducts or tube-like extensions from the surface (Fig. 20.2e ) (Montgomery and McFall-Ngai 1993). Passage through the ducts appears to be a challenge: although heterologous species such as V. parahaemolyticus appear competent to reach the duct, they fail to colonize (Nyholm et al. 2000). The duct contains mucus, cilia that appear to beat outward toward the pores, and antimicrobial molecules (Davidson et al. 2004; McFall-Ngai and Ruby 1998; McFall-Ngai 1999; Small and McFall-Ngai 1999). To progress to colonization, V. fischeri must be able to overcome these challenges and others, as described in greater detail below.
Each of the six ducts leads to an antechamber (Sycuro et al. 2006), a small chamber outside of the larger deep crypt where most colonizing cells eventually reside (Fig. 20.2e ). Each set of antechambers has an average size and complexity that corresponds to that of the deep crypt with which it is associated. For example, the antechamber of crypt 1 has an average cross-sectional area of 1,380 μm2, while the antechamber of crypt 3 has its largest cross-sectional dimension in the range of 510 μm2 (Sycuro et al. 2006) (Fig. 20.2e ). The antechambers are not permissive to persistent colonization, likely because this is a region of the light organ with extensive antimicrobial activities, such as nitric oxide (NO) production (Davidson et al. 2004). Little else is known about the antechambers. To reach their respective deep crypts, the bacteria must exit the antechamber through a bottleneck region that has small dimensions (between 5 and 9 μM in width) that limit passage.
Like the antechambers, the three sets of deep crypts have different characteristics with respect to size and complexity. In the newly hatched juvenile, the largest and most developed is known as deep crypt 1 (or simply crypt 1), while the smallest and least developed is crypt 3. Crypt 2 is intermediate between the other two. Because of the complexity of the deep crypt tissues, no estimate has been made of the area or volume of these spaces. Each of the deep crypts in the uncolonized juvenile is lined with columnar epithelial cells. The microvilli on the surfaces of these cells in colonized animals provide points of direct contact with the bacterial symbionts (Fig. 20.2f ). It is within these confined deep crypt spaces that multiplication to high cell density ensues, resulting in colonization of the host by V. fischeri. Sycuro et al. (2006) reported that there is no particular order to which the six deep crypts become colonized. Similarly, Dunn et al. (2006) noted that, while bacterial gene expression was altered in crypt 3 relative to the other crypts, the timing of colonization did not appear different. Thus, there appear to be differences in crypt structure and maturity in hatchlings, but the differences do not interfere with colonization. Rapid growth of the bacteria is supported by host-provided nutrients, including amino acids presented in the form of peptides (Graf and Ruby 1998), and likely oxygen. When a sufficiently high cell density is achieved, the production of bioluminescence is induced (Ruby and Asato 1993).
It is readily apparent from this brief description that V. fischeri experiences a variety of environments during its passage into the deep crypts where multiplication and colonization occurs. V. fischeri must transit from the nutrient-limited seawater to the mucus-lined surfaces of the light organ, through the apparently challenging environments of the ducts and antechambers, to reach the nutrient-rich, hospitable environment of the deep crypts where rapid growth is possible and a generation time as low as 30 min is estimated (Ruby and Asato 1993). Each stage likely requires the expression of a distinct set of traits that permit V. fischeri—and no other bacteria—to successfully navigate these challenges.
Dynamics of Symbiosis
One of the most interesting facets of the V. fischeri-squid symbiosis is its dynamic nature. The squid are nocturnal animals, and many of their behaviors are cued to the day/night cycle. For example, the animals forage for food at night, but bury in the sand during the day (Fig. 20.3 ) (Moynihan 1983). In addition, juveniles hatch from eggs at dusk; thus, these newly hatched squid become colonized at night. That event begins a cycle of colonization, expulsion, and regrowth: every dawn, colonized squid expel 90% of their bacterial symbionts by means of a muscle-induced contraction (Fig. 20.3 ) (Boettcher et al. 1996; Graf and Ruby 1998; Lee and Ruby 1994b; Nyholm and McFall-Ngai 1998; Ruby and Asato 1993). In the adult, the result is the release of a toothpaste-like gel of acellular matrix along with bacteria and host cells (Nyholm and McFall-Ngai 1998). The remaining 5–10% of the bacterial population repopulates the light organ. The consequence of this phenomenon is that both the bacteria and their host experience a changing environment daily. In this section, we describe some of the known molecular details that correspond to the rhythm of the squid’s biology, including the early events specifying colonization and subsequent daily events that influence the interaction between the partners.
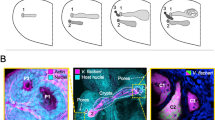
Detailed studies of the initiation of colonization revealed a surprising fact: within the first hour following hatching (in fact, as early as 20 min), the light organ is permissive to entry by both V. fischeri and non-V. fischeri bacteria, as well as other similarly sized particles (Fig. 20.4a ) (Nyholm et al. 2002). Both Gram-negative and Gram-positive bacteria with sizes of 1 μm in diameter could be observed in the crypt spaces in the first hour. Fluorescent beads with a 1-μm diameter could also be observed in the deep crypts, but not beads with larger diameters (2 μm or 10 μm) or cells (Bacillus cereus) with a diameter of 5 μm. This phenomenon was labeled the permissive period, since even nonsymbiotic bacteria can gain entry. However, no viable bacteria could be recovered from light organs at this time using plating techniques, and furthermore, no particles (bacteria or beads) could be detected 2 h after inoculation (Nyholm et al. 2002). Thus, following entry, the bacteria and beads appear to be removed, likely by host defense cells (hemocytes) (Nyholm et al. 2009), and the time between 1 and 2 h after hatching represents a nonpermissive period.
Initiation of colonization. (a) An approximate time line of some of the known events in colonization during the first 48 h is depicted. Following hatching (0 h), the light organ is transiently permissive to entry by bacteria (0.5 h). Mucus shedding (represented by fuzzy shading on the surface of the organ) is induced (2 h), promoting surface aggregation by V. fischeri (ovals) (3 h). Colonization occurs (darker shading) (12 h) and triggers apoptosis (represented by dots), regression of the appendages, and cessation of mucus secretion (48 h). (b) Images of light organs clonally or dually colonized by GFP- or RFP-expressing V. fischeri cells. The segregation of initiating cells is apparent by the separation of colors; in the last panel, mixing of cells can be seen (white arrow) (Images from panel b were previously published Dunn et al. (2006))
Within 1–2 h following hatching, mucus secretion occurs from cells within the ciliated epithelial appendages in animals exposed to bacteria, but not in response to beads (Fig. 20.4a ) (Nyholm et al. 2002). Sialomucin is the predominant mucin type found on the surface of the epithelial fields, but neutral mucins can also be detected. Although sialomucin can be found on the surfaces of the appendages of squid maintained in filter-sterilized seawater, no shedding of mucus occurs. Mucus shedding could be induced by the addition of peptidoglycan (PG) but not lipopolysaccharide (LPS) (Nyholm et al. 2002). Mucus secretion also could be induced by PG-coated beads too large to enter the light organ, indicating that entry is not necessary and that a receptor for PG must be present on the surface of the light organ. These data indicate that the early permissive period is not essential for induction of mucus secretion, if PG is present in the seawater. In contrast to several developmental events discussed below that require V. fischeri and also involve PG, this induction of mucus secretion occurs even if V. fischeri is absent from the seawater, indicating that the squid are simply sensing the presence of bacteria.
Because the onset of mucus secretion coincides with the beginning of the nonpermissive period, experiments were undertaken to determine if mucus secretion causes the block to particle entry. Newly hatched juvenile squid were exposed to PG for 3 h to induce mucus secretion. These animals were then exposed to GFP-labeled nonsymbiont V. parahaemolyticus, and the number of animals with these bacteria in their crypts determined. The results indicated that reduced numbers of animals with bacteria could be detected relative to animals that had not been exposed to PG and thus not shedding mucus. The authors of this study concluded that mucus secretion contributes to, but is not wholly responsible for, the block to the permissive period (Nyholm et al. 2002).
The production of mucus promotes the ability of V. fischeri to aggregate on the surface of the light organ and subsequently to enter and colonize (Fig. 20.4a ). Once V. fischeri has colonized, however, mucus shedding is downregulated: the amount of mucus secreted from 72-h aposymbiotic animals was significantly greater than that of 48-h colonized animals (Nyholm et al. 2002). Furthermore, V. fischeri could aggregate on the light organs of aposymbiotic animals upon exposure at any point during the first 4 days, but could not aggregate on the light organs of 48-h symbiotic animals, which shed relatively little mucus (Fig. 20.4a ). Presumably, once colonization is achieved, there is no longer a need for mucus to promote bacterial aggregation, and thus it is downregulated; this decrease in mucus production likely also restricts colonization by undesired species. In further support of the relationship between colonization and mucus shedding, symbiotic animals that were cured of their bacteria through antibiotic treatment exhibited an increase in mucus shedding, which once again promoted V. fischeri aggregation. It should be noted that cured animals, while able to shed more mucus and permit aggregation, exhibited reduced levels of aggregation, likely due to the reduced numbers of mucus-secreting epithelial cells resulting from apoptosis and regression of the ciliated appendages (Fig. 20.4a ), developmental events that occur in the same time frame (described below) (Nyholm et al. 2002). Finally, the deep crypt spaces of colonized animals contained increased mucus compared to uncolonized animals (Nyholm et al. 2002). This effect was opposite to the downregulation that occurs on the light-organ surface, indicating that mucus secretion in the crypts may promote symbiosis.
V. fischeri also downregulates the production by the host of nitric oxide (NO), which is relatively high in uncolonized hatchlings (Davidson et al. 2004). NO synthesis carries both a cost to produce and a risk (of oxidative damage) to the host. By shutting down mucus and regressing the appendages, the squid has greatly reduced the opportunity for further colonization and, potentially, can now relax its defenses. Sustained activation might ultimately jeopardize maintenance of symbiont colonization, and thus, this change in NO may reflect an accommodation for V. fischeri. Alternatively, it might reflect a signaling function for NO (see below). Regardless, it is clear that the result of colonization is to decrease subsequent attachment and superinfection. These phenotypes demonstrate the influence of V. fischeri on the biology of its host.
This tight control over initiation of symbiosis begs the questions, how many cells are necessary to initiate colonization and how many different V. fischeri cells can successfully colonize the light organ of a single squid? These questions were experimentally addressed in a series of studies. Early studies indicated that squid could contain more than one strain. For example, bacteria with different plasmid profiles could be isolated from the light organ of a single field-caught adult animal (Boettcher and Ruby 1994). In addition, when a mixture of two strains was used to inoculate juvenile squid, some colonized animals contained both strains, indicating that multiple strains could colonize (Lee and Ruby 1994a). Furthermore, a marked strain could be introduced into a colonized animal, albeit at a very low frequency (Lee and Ruby 1994a); likely the poor efficiency of superinfection resulted from decreased mucus shedding and loss of the ciliated surface appendages.
A subsequent study evaluated how different numbers of bacteria impacted colonization proficiency. Generally, at different inoculation dosages, the percentage of squid that had associated bacteria at an early time point (3 h) was similar to the percentage of squid that ultimately became colonized (McCann et al. 2003). Inoculation with as few as 250 V. fischeri cells in 4 ml during a short (3-h) period of time was sufficient to promote colonization of 50% of the animals, indicating that colonization by V. fischeri is an efficient process (McCann et al. 2003). At inoculation levels above 1,000 bacteria, colonization occurred 100% of the time. With increasing doses of bacteria, the efficiency of colonization increased, as determined by the decreasing time to onset of luminescence, a trait that is governed by cell density. However, beyond a certain point, no further increase in efficiency was obtained with increasing numbers, suggesting that the process rather than the number of bacteria becomes limiting.
The same study asked whether three strains of V. fischeri could simultaneously colonize a single squid (McCann et al. 2003). The strains differed only by distinct antibiotic markers that did not substantially impact colonization proficiency of a single strain. At a low inoculation dose (500 cells), all colonized squid contained a single strain. When the dosage was increased to 5,000 cells, most squid contained two strains but not three. Finally, at high doses (16,000 and 27,000), a significant percentage of squid contained all three strains. Thus, although multiple strains can co-colonize, it appears that, at levels similar to those found in nature (Lee and Ruby 1992), only one or two bacteria generally colonize a single animal.
A subsequent study evaluated co-colonization using two strains that differed by a fluorescent tag (red or green fluorescent protein) and visual examination of crypt colonization using epifluorescence microscopy (Dunn et al. 2006). When squid were inoculated with moderate doses of the two strains (for a total of 2,000–7,000 CFU/ml), most animals were colonized with both strains. Interestingly, however, most animals with mixed infections contained pockets of either red or green fluorescence, and only rarely did light organs contain a region with a mixture of red and green cells (Fig. 20.4b ). These data further support the idea that a few cells initiate colonization, and even when multiple strains colonize, they often become segregated within the light organ, presumably because it is often only a single cell that initially reaches each deep crypt and becomes the dominant colonist there.
These two experimental studies, which suggest that low numbers of V. fischeri cells initiate colonization, are consistent with a subsequent analysis examining the population structure of V. fischeri. Wollenberg and Ruby (2009) used an extensive PCR-based analysis along with phenotypic analyses and modeling to evaluate the number of different strains of V. fischeri in light organs of field-caught animals. These analyses support the prediction that one or two bacteria colonize each crypt, resulting in the mixed population structure found within the adult light organ (Wollenberg and Ruby 2009).
As mentioned earlier, E. scolopes expel ~90% of their bacterial symbionts every dawn (Boettcher et al. 1996; Graf and Ruby 1998; Lee and Ruby 1994b; Ruby and Asato 1993). Intriguingly, for juveniles, different crypts are emptied with different efficiencies: bacteria are expelled from deep crypt 1 to a greater extent than crypt 2, with crypt 3 being least effectively emptied (Sycuro et al. 2006). Sycuro et al. (2006) suggest two possibilities for these results: the expulsion efficiency (1) reflects the relative developmental maturity of the crypts or (2) is due to position effects (the muscle contraction is less effective in the area where crypt 3 resides). These features of the juvenile could impact the identity and number of specific symbiont strains over time. Indeed, one study determined that a specific mutant strain of V. fischeri was preferentially expelled from the light organ in mixed colonization experiments (Millikan and Ruby 2004). Understanding the dynamics of crypt expulsion and how it relates to colonization competitiveness and persistence is an important area of future investigation.
Linked to but separate from the expulsion event is another rhythm established in E. scolopes, the variation of bacterial bioluminescence over the day/night cycle. Luminescence increases and is highest in the hours preceding darkness; light production is as much as 100-fold lower at other times (Boettcher et al. 1996). This rhythm of increasing and decreasing light emission is disrupted if animals are kept in either constant light or constant darkness, indicating that it is a diel rhythm rather than a circadian rhythm (Boettcher et al. 1996). Furthermore, the amount of specific bioluminescence, or the amount of light produced on a per-cell basis, varies over the day/night cycle: when symbiotic luminescence levels are high prior to the onset of darkness, the amount of symbiotic light per cell matches that produced by newly released bacteria (Boettcher et al. 1996). However, in periods of low symbiotic luminescence, the specific luminescence is lower in symbiotic cells relative to newly released cells. These data indicate that the squid controls (inhibits) light production or emission. The daily expulsion of bacteria at dawn contributes to the change in light emission over the daily cycle: a peak of luminescence can be observed at the transition from dark to light that is not observed for animals kept in constant light or dark conditions. The peak of light corresponds to the release of bacteria into the seawater; following this expulsion event, the levels of luminescence by the squid are clearly decreased. However, although expulsion clearly contributes to a drop in light intensity, it is insufficient to fully account for the observed patterns, as light emission decreases steadily over a number of hours prior to the expulsion event.
Another mechanism by which the squid controls light emission is by physically blocking it. It is clear that adult animals can direct, control, and conceal the light produced by their bacterial partner using a muscle-controlled ink sac, along with lens and reflector tissues (McFall-Ngai and Montgomery 1990). These tissues direct the light downward to match the downwelling moonlight and modulate light emission under a variety of natural environmental conditions (e.g., full moon and new moon), thus permitting the squid to use counterillumination as a defense mechanism (Jones and Nishiguchi 2004). However, the light organs of newly hatched juvenile squid do not seem to have sufficiently developed accessory tissues to account for the observed diel rhythm (Boettcher et al. 1996; McFall-Ngai and Montgomery 1990; Montgomery and McFall-Ngai 1993). Thus, it was speculated that another mechanism must be in place to account for the daily modulation of bioluminescence.
The currently favored model is that the level of specific luminescence varies over the daily cycle due to changes in oxygen levels provided to the symbiont by the host (Boettcher et al. 1996). Luminescence is an oxygen-dependent reaction, and thus decreased oxygen availability could lead to decreased luminescence. The finding that newly released bacteria exhibit a spike in their levels of light production is consistent with the idea that oxygen is more readily available in seawater and, as a limiting reagent in the light organ, can rapidly change the specific luminescence level. Regardless of the cause, changes in luminescence are part of the daily rhythm of the symbiosis.
A series of studies has investigated the influence of V. fischeri and the daily rhythm on host transcription and protein production. One of these studies generated a set of 11 expressed tag sequence (EST) cDNA libraries from juvenile animals at different times and either with or without V. fischeri cells (colonized or uncolonized) (Chun et al. 2006). A large number (46%) of the non-redundant set of sequences appeared to represent unique sequences, as they had no matches in the database at the time. Perhaps the most significant finding from the initial description of this tool was that the biggest set of unique transcript fragments was obtained from animals that had been colonized for 2 days (48 h). This finding agrees with previous proteomic work showing that 48 h was the earliest time point at which differences in protein profiles between colonized and uncolonized animals could be detected (Doino Lemus and McFall-Ngai 2000). At this point in colonization, numerous developmental changes are occurring or are being induced by the bacteria, including apoptosis and regression, and crypt cell swelling (Nyholm and McFall-Ngai 2004). The proteomic study also found that some proteins that appeared specifically in 48-h symbiotic animals but not in uncolonized animals were not symbiosis-specific, as the same proteins could be detected in uncolonized animals at a later time point (96 h) (Doino Lemus and McFall-Ngai 2000). These data indicate that the bacteria accelerate certain host developmental events in addition to inducing symbiosis-specific gene expression.
The EST database permitted the construction of a microarray, which was used to probe changes in host gene expression in response to the symbiosis. Specifically, a subsequent study investigated how host transcription was affected by colonization, luminescence, and the LuxI-produced autoinducer pheromone that induces luminescence (as described in greater detail below) (Chun et al. 2008). Perhaps not surprisingly, the biggest influence on the response of the host was the presence of the bacteria, with hundreds of transcripts altered in response to colonization. The host also responded to light production and, to a much lesser extent, the presence of autoinducer. Further investigation of the transcriptional responses and the impact of those changes on host activities will provide important insights into host colonization.
A second microarray study analyzed gene expression of both V. fischeri and its host at 6-h intervals to capture the changes that drive or derive from the squid’s daily rhythm (Wier et al. 2010). For the host, about 10% of genes present in the array (a ~14,000 gene EST library) exhibited changes in expression. The 6-h intervals on either side of dawn showed the greatest differences in host gene expression, notably including changes in transcripts encoding proteins with cytoskeletal functions. In the interval prior to dawn, there was a substantial upregulation in expression of this suite of genes, while in the period after dawn, they were substantially downregulated, suggesting cytoskeletal remodeling was induced then decreased in the intervals bracketing dawn. An examination of the crypt epithelium revealed that structural changes indeed occurred right around dawn (Wier et al. 2010). Throughout most of the 24-h period, the host epithelial cells appear “normal”: the cells are highly polarized and have microvilli on the surfaces that interact with the symbionts. However, around dawn, the cell surfaces appear effaced, and portions appear to be released into the crypts as vesicles (Nyholm and McFall-Ngai 1998; Wier et al. 2010). Within a few hours, however, the normal appearance is restored. Thus, there is a daily cycle of structural change in the host epithelium coincident with symbiont expulsion.
The same microarray experiment confirmed that the symbionts also changed their gene expression: 17% of genes changed during one of the four intervals, with the greatest period of regulatory change occurring in the interval after dawn (Wier et al. 2010). A deeper analysis indicated that genes involved in the catabolism of chitin were upregulated in the period prior to dawn and downregulated after dawn and throughout the day. In contrast, genes involved in glycerol metabolism were upregulated after dawn. Wier et al. (2010) proposed that host vesicles, which become abundant around dawn, would be a rich source of glycerophospholipids, from which glycerol could be released (Wier et al. 2010). In support of the idea that the symbionts could incorporate fatty acids from host vesicles into their membranes, symbiont cells contained dramatically different lipid profiles compared to cultured V. fischeri (Wier et al. 2010). Together, these data suggest that the metabolism of V. fischeri undergoes dynamic changes over the course of each day, depending on the availability of nutrients.
In addition to light production and, likely, metabolism, V. fischeri undergoes developmental changes that are reflected in the daily rhythm. Motility, which is necessary for V. fischeri to enter and reach the deep crypts, appears unnecessary upon colonization as deep-crypt-localized V. fischeri have no flagella (Ruby and Asato 1993). In addition, V. fischeri cells undergo a change in morphology: within 24 h of symbiosis, the cells have become smaller and rounder than culture-grown cells or symbionts within the first 12 h of symbiosis (Ruby and Asato 1993). The signals and genes responsible for inducing these changes remain unknown.
In summary, the light-organ symbiosis is dynamic. The bacteria and their host influence each other’s gene expression and induce developmental events, promoting, among other things, an exquisite specificity in partner selection and reducing competition by others, including late-arriving V. fischeri. For the host, some changes, once initiated, are irreversible, while others require the continuous presence of the bacteria. Numerous events occur on a daily cycle driven by a number of factors, including bacterial expulsion, available nutrients and (probably) available oxygen, and bacterial growth. This dynamic nature must be considered when assessing the requirements for specific bacterial traits, which may be important at one stage or one temporal period, but not another. This is one example where the laboratory “batch culture” may actually somewhat reflect a natural process in the wild.
Specificity and Host Defenses
The discrete localization of the V. fischeri light-organ infection and the inability of other bacteria to colonize this tissue demonstrate an exquisite level of control by the host. As further evidence that host tolerance of the symbiont is regulated, we have observed that when juvenile squid that are colonized by V. fischeri become nutritionally stressed, the animals are able to entirely clear the V. fischeri infection (Stabb, unpublished data). Thus, while the light organ is receptive to infection and able to support the rapid growth of V. fischeri, the squid are able both to prevent other bacteria from colonizing and to keep the V. fischeri symbionts themselves in check. The mechanisms of specificity and control of the infection maintained by the host remain intriguing and somewhat mysterious, although a great deal is now known. It appears that specificity is achieved through multiple layers of enrichment, eventually “winnowing” V. fischeri symbionts away from unwanted interlopers (Nyholm and McFall-Ngai 2004). The underlying mechanisms involve physical barriers, physiological constraints, and immune functions that include both broad-spectrum antimicrobial compounds and potentially a more microbe-specific population of macrophage-like hemocyte cells (Fig. 20.5 ).
Cilia and mucus in the ducts may constitute an impediment to infection (Nyholm and McFall-Ngai 1998). A sticky mucus also facilitates adherence to the outside of the light organ by planktonic cells, but such adherence could constitute a barrier to the movement required to traverse the ducts. Once the symbionts are inside the crypts, junctions between epithelial cells probably form a physical barrier that keeps them constrained to the light organ (Nyholm and McFall-Ngai 1998). Although the V. fischeri genome appears to encode a Zot toxin that theoretically could disrupt tight junctions (Ruby et al. 2005), gfp-labeled V. fischeri have not been observed escaping the epithelium-lined crypts of the light organ. The squid’s active daily expulsion of most light-organ lumen contents provides a physical limitation on bacterial overgrowth otherwise breaching the crypt epithelium, and it would presumably also enrich for bacteria that have the ability to avoid being expelled. As described below, V. fischeri appears to have a variety of pili and adhesins that may have coevolved with the host for this purpose (Browne-Silva and Nishiguchi 2008; Ruby et al. 2005).
The daily cycle of regrowth of bacteria in the light organ may give the host an additional mechanism for maintaining the specificity of its symbionts. For example, by providing or withholding certain nutrients, the growth of V. fischeri may be favored over other bacteria that gain access to the light organ. Such a mechanism remains speculative, and it would be difficult if not impossible to test the growth rate of different bacterial species within the physiological parameters of the light organ in the absence of other (e.g., immunological) specificity factors. It seems unlikely, however, that this could be the major factor in maintenance of specificity: the contents of the light organ appear complex and include substrates (e.g., peptides and amino acids) that could support the growth of numerous other bacteria. Although iron appears to be limiting, and iron-uptake machinery may be an important colonization factor for V. fischeri, many other marine bacteria have similar iron-scavenging systems. Thus, nutrient control is likely, at best, a mechanism to enrich for ongoing V. fischeri colonization.
Instead, it seems likely that the major control over the symbiont population and specificity is exerted by the host immune system. Naturally, this topic has been an area of great research interest, and the role of the E. scolopes innate immune system in the symbiosis was recently reviewed (McFall-Ngai et al. 2010). Discovery and elucidation of E. scolopes immune functions have been accelerated by the generation of EST libraries (described above) that profile the host transcriptome (Chun et al. 2008) and the characterization of the host light-organ proteome (Schleicher and Nyholm 2011). For example, analysis of ESTs led to the discovery of components of an immunological complement system, followed by experimental determination that complement C3 protein is expressed on the apical surface of light-organ epithelial cells (Castillo et al. 2009). Research can now address whether the complement system helps direct immunological responses that maintain specificity or constrain V. fischeri. There is also evidence that the squid are capable of producing antimicrobial peptides (Nyholm and McFall-Ngai 2004), which may have an immune function that is either broad-spectrum or weighted toward the control of non-V. fischeri.
Among the potential antimicrobial components of the squid innate immune response, the most thoroughly studied to date have been reactive oxygen species (ROS). Recent analysis of the host proteome suggested a number of highly expressed proteins are involved in producing ROS (Schleicher and Nyholm 2011). For example, the host apparently encodes a number of putative ROS-generating peroxidases, including the halide peroxidase (HPO) described below. In the same study, it was found that symbiotic host and V. fischeri cells contained numerous putative antioxidant proteins. For example, one of the most abundant proteins in symbiotic V. fischeri cells was AhpC, a predicted alkyl hydroperoxide reductase (Schleicher and Nyholm 2011). These recent data are consistent with the longstanding hypothesis that oxidative stress is a hallmark of the light-organ environment, and they provide new targets for future investigations.
Interest in E. scolopes ROS production began with the discovery that the squid expresses HPO in the light organ, presumably producing the antimicrobial ROS hypochlorous acid (HOCL). Peroxidase-encoding transcripts were among the most abundant found in early cDNA libraries from E. scolopes, and the HPO transcript was among the first expressed genes discovered from the light organ (Tomarev et al. 1993). Further studies confirmed the biochemical similarity of the squid-encoded halide peroxidase to mammalian peroxidases, including production of hypohalous acid from halide ions and H2O2, as well as its presence in the light organ (Weis et al. 1996; Small and McFall-Ngai 1999). Given the high level of chloride ions in seawater, it seems likely that the main relevant product of HPO is HOCl, an effective broad-spectrum antimicrobial used commercially as a disinfectant. The HPO gene in E. scolopes is more highly expressed in tissues that are exposed to bacteria, including gills as well as the light organ (Small and McFall-Ngai 1999). Like other bacteria, V. fischeri is sensitive to HOCl, suggesting that HPO represents a broad-spectrum antimicrobial that keeps bacteria in check rather than a light-organ-specific mechanism for selecting V. fischeri. Interestingly, recent studies show that HPO is localized in the host hemocyte cells (Heath-Heckman and McFall-Ngai 2011).
Given that H2O2 is a substrate for HPO, it seems likely that this ROS is produced in the light organ as well, perhaps via a host respiratory burst. Like many bacteria, V. fischeri encodes a catalase (KatA) that converts H2O2 to water and oxygen, but the periplasmic location of V. fischeri is unusual and suggests a key role in detoxifying H2O2 originating from an external source (Visick and Ruby 1998). The relatively high catalase activity in V. fischeri likewise points to a role beyond coping with endogenous metabolic production of H2O2 (Visick and Ruby 1998). A V. fischeri katA mutant was able to colonize the E. scolopes light organ to wild-type levels when presented as a clonal inoculum but was outcompeted by the wild type in mixed competitive infections, indicating that catalase contributes to, but is not required for, colonization of the host (Visick and Ruby 1998). Analysis of the V. fischeri genome suggests that while katA encodes the bacterium’s only catalase, there are other mechanisms for coping with oxidative damage. For example, there are three methionine sulfoxide reductase genes that presumably repair proteins damaged by H2O2 (Flores and Stabb, unpublished data). Redundancy in the V. fischeri response to this and other ROS may explain why mutants lacking single oxidative-response enzymes are not more severely attenuated in colonization.
Another oxidant produced by E. scolopes is nitric oxide (NO). Both NO and NO synthases (NOS) are produced throughout light-organ tissues, and both are found in mucus secretions that contact V. fischeri symbionts (Davidson et al. 2004). Although both V. fischeri and other species aggregate in host-derived mucus on the surface of the light organ, V. fischeri is somehow enriched (Nyholm et al. 2000, 2002; Nyholm and McFall-Ngai 2003). NO-scavenging molecules increased the number of either V. fischeri or nonsymbionts in such aggregates on the light-organ surface (Davidson et al. 2004). Although nonnative bacteria were still unable to colonize the light-organ crypts, the results suggest that NO limits bacterial growth in the aggregates and could play some role in the enrichment seen at this stage (Davidson et al. 2004).
The transcriptional response of V. fischeri to NO was recently elucidated (Wang et al. 2010a), leading to intriguing discoveries and new research directions. For example, a heme-independent and NO-resistant alternative oxidase gene (aox) is controlled by the NO-responsive regulator NsrR and is upregulated in response to NO (Dunn et al. 2010). Aox allows aerobic respiration to continue when other oxidases in the cell are inhibited by NO, which may impart a competitive advantage to V. fischeri over the majority of other Vibrio species, which lack aox (Dunn et al. 2010; Spiro 2010). NO production also results in the H-NOX-dependent downregulation of iron acquisition (Wang et al. 2010a). While this response may not relate directly to NO resistance, it could reflect NO being used by V. fischeri as an indicator of other ROS it will soon encounter, specifically H2O2. By limiting iron uptake, symbionts might limit the oxidative stress generated by Fenton chemistry when H2O2 and iron are combined (Wang et al. 2010a). Before leaving the topic of NO, it should be noted that it may play an additional symbiotic role in addition to functioning as an antimicrobial oxidant: NO has signaling functions in many higher organisms, and as described below, it may be part of the morphogenic developmental program in the light organ stimulated by V. fischeri (Altura et al. 2011).
In addition to producing broadly antimicrobial molecules, E. scolopes has a cellular immune response involving hemocyte cells (Fig. 20.5 ). These immune cells may play a key role in maintaining the specificity of the interaction with V. fischeri (Nyholm and McFall-Ngai 1998; Nyholm et al. 2009). As is the case in other cephalopods, E. scolopes appears to have a single type of hemocyte, which circulates in the blood and moves throughout the animal. Like mammalian macrophages, these hemocytes can bind, engulf, and kill bacterial cells. E. scolopes hemocytes have been found in the blood as well as in the light-organ crypts and the sinuses of the ciliated epithelial appendages (Fig. 20.2c ) (Koropatnick et al. 2004; Nyholm and McFall-Ngai 1998). Within the light-organ crypts of newly hatched juveniles, the hemocytes have been observed with internal bacteria, presumably engulfed; however, in adult animals, the hemocytes were seen surrounded by densely packed V. fischeri cells but had not phagocytosed them (Nyholm and McFall-Ngai 1998; Nyholm et al. 2009). These data suggested that the hemocyte cells can ignore V. fischeri and that this may be a trait acquired as the animals develop. When removed from the squid, the macrophage-like hemocytes bound and phagocytosed V. fischeri less frequently than they did other marine bacteria (Nyholm et al. 2009). Binding seemed to be the key rate-limiting step in this process, as bound V. fischeri were as likely to be phagocytosed as another bacterium (Nyholm et al. 2009).
In an interesting twist, Nyholm et al. (2009) also found that exposure to V. fischeri was critical for maintaining hemocyte specificity (Nyholm et al. 2009). When squid were cured of their V. fischeri symbionts with antibiotics, hemocytes from these symbiont-free animals became five times more effective at binding V. fischeri while their affinity for V. harveyi or Vibrio parahaemolyticus was unchanged (Nyholm et al. 2009). Moreover, hemocyte binding to V. fischeri was similarly high in hemocytes isolated from colonized or naïve animals when the target V. fischeri strain was a mutant lacking the major outer membrane protein OmpU (Nyholm et al. 2009). These data suggest that E. scolopes hemocytes learn to discriminate V. fischeri from other bacteria and adapt to preferentially bind nonsymbionts through an OmpU-dependent mechanism. Interestingly, it was recently reported that Vibrio splendidus OmpU mediates adhesion to and invasion of oyster hemocytes (Duperthuy et al. 2011). While the two OmpU-mediated phenomena seem opposite to each other, the underlying processes involved may reveal quite parallel mechanisms once they are understood.
Another observation of E. scolopes hemocytes that may have widespread importance is the discovery of chitin and endogenous chitin synthesis within these immune cells (Heath-Heckman and McFall-Ngai 2011). This was found to be a common property of invertebrate hemocytes that is lacking in their vertebrate counterparts (Heath-Heckman and McFall-Ngai 2011). V. fischeri can metabolize chitin and ferment its N-acetylglucosamine monomers, but it remains to be determined what, if any, role hemocyte-derived chitin plays in E. scolopes immunity or support of V. fischeri growth. From multiple perspectives, the biology of E. scolopes hemocytes appears worthy of further investigation likely to reveal elements unique to the V. fischeri-E. scolopes symbiosis as well as phenomena more broadly applicable to invertebrate-bacteria interactions.
Host Development and Bacterial Signals
Host Development
V. fischeri is the lone bacterial species colonizing the E. scolopes light organ, and infection by V. fischeri is required to trigger developmental changes in the host, some of which can be mimicked using bacterially derived molecules. If kept in water free of V. fischeri, E. scolopes can be raised to adulthood without the light organ becoming infected or bioluminescent (Hanlon et al. 1997). Such aposymbiotic animals are healthy and develop normally in most respects, including the lens and reflective tissue of the light organ (Claes and Dunlap 2000). However, specific developmental events fail to occur in the absence of V. fischeri infection, including, most dramatically, the regression of the ciliated fields on the light organ. At the time of hatching, the ciliated fields begin shedding mucus, which as described above helps facilitate infection; however, once the light organ is infected, mucus shedding ceases (Nyholm et al. 2002), and this cessation is followed by regression of the structures themselves. Over the course of 4–5 days postinfection, these infection-promoting structures are completely lost in infected animals (Fig. 20.6 ) (Montgomery and McFall-Ngai 1994; Foster and McFall-Ngai 1998; McFall-Ngai and Ruby 1991). This morphological change is accompanied by apoptosis in the epithelial cells of the ciliated fields and infiltration of host hemocytes into the sinuses of the ciliated appendages (Koropatnick et al. 2004, 2007). None of these developmental events take place in aposymbiotic animals.
Development of the juvenile light organ. Epifluorescence (Epi) and scanning electron microscopy (SEM) images of the stages of regression of ciliated epithelial appendages. Upon exposure to peptidoglycan or symbiosis-competent V. fischeri, the ciliated appendages present on newly hatched squid (stage 0) undergo thinning (stage 1) and progressive shortening (stages 2 and 3), until they are lost (stage 4, not shown). aa anterior appendage, pa posterior appendage, p pore, and cr ciliated ridge (These images were previously published Adin et al. (2009))
V. fischeri also triggers more subtle developmental effects in the light organ. For example, the ducts leading from the light-organ surface to the crypts constrict (Kimbell and McFall-Ngai 2004), and the cells lining the duct become more homogenous and filled with inclusions (Claes and Dunlap 2000). The epithelial cells lining the crypts swell (Montgomery and McFall-Ngai 1994) with a proliferation of microvilli on their surfaces (Lamarcq and McFall-Ngai 1998), and there is an apparent increase in mucus secretion inside the crypts themselves (Nyholm et al. 2002). Additional V. fischeri-dependent molecular events, including a downregulation of NO synthase (Davidson et al. 2004), have been observed but not yet clearly linked to developmental and physiological processes (Doino Lemus and McFall-Ngai 2000; Kimbell and McFall-Ngai 2003; Chun et al. 2008). Many of these developmental events triggered by V. fischeri, as well as their timing, were reviewed by Nyholm and McFall-Ngai (Nyholm and McFall-Ngai 2004).
In general, most symbiont-induced developmental changes in the E. scolopes light organ can be rationalized in terms of this organ’s two temporally distinct functions—first, to become infected with V. fischeri and then later to support and control symbiotic bioluminescence. For newly hatched aposymbiotic squid, acquiring symbionts from a dilute environment is a numerically daunting task that is facilitated by the ciliated cells and the mucus they secrete (Nyholm et al. 2000, 2002). However, once the squid are colonized by appropriate V. fischeri symbionts, the infection-promoting properties of the ciliated fields become unnecessary and could perhaps be a liability if pathogenic infections were facilitated. Thus, programmed cell death and regression of the ciliated fields and constriction of the ducts may serve to prevent further infection beyond the initial colonization with a mutualistic V. fischeri symbiont. The developmental events inside the crypts, cell swelling and microvillar proliferation, may serve to increase the surface area at the symbiont-host interface and promote the efficient exchange of metabolites.
The symbiont-triggered developmental events in E. scolopes appear to result from multiple distinct signaling pathways. Some changes are reversible if E. scolopes is cured of its symbionts with antibiotics (Lamarcq and McFall-Ngai 1998; Nyholm et al. 2002), whereas other events cannot be stopped once they are set in motion (Doino and McFall-Ngai 1995). For example, the swelling of crypt epithelial cells can be reversed by curing the symbionts, and microvillar proliferation on these cells does not progress and may even reverse somewhat if symbionts are cured (Lamarcq and McFall-Ngai 1998). In contrast, regression of the ciliated epithelial fields does not require persistent colonization after about 12 h postinoculation (Doino and McFall-Ngai 1995). Regression proceeds in animals that have been cured of symbionts, and the ciliated epithelial structures do not grow back. Thus, it seems that the programmed loss of infection-promoting structures is set in motion early during infection and does not require constant colonization. In one exception to this, the infection-promoting mucus secretion of the ciliated structures does reappear in animals that are infected and then cured of V. fischeri (Nyholm et al. 2002).
Taken together, the results of several early experiments indicate a complex pattern of light-organ development that includes symbiont-dependent and symbiont-independent programs, some of which require only transient exposure to V. fischeri. Moreover, although Claes and Dunlap (2000) noted that the tissues developmentally affected by V. fischeri all come in contact with symbiotic cells, at least some developmental events appear to involve symbiont induction remotely. Specifically, the ciliated fields on the light-organ surface are exposed to V. fischeri, but mutants unable to colonize the crypts still contact these cells on the surface of the light organ without triggering their regression. Once the crypts become packed with V. fischeri cells, regression of the ciliated appendages advances, even though symbionts in the crypts are several cell layers away.
Bacterial Signals That Influence Host Development
The observation that infection with V. fischeri triggers developmental programs and morphological changes in the E. scolopes light organ has prompted interest in understanding the specific mechanisms and bacterial signals involved in these processes. Most research has focused on the involvement of three bacterial factors in stimulating host development: bioluminescence, LPS, and PG. Both LPS and PG can be categorized as microbe-associated molecular patterns (MAMPs), and they have intriguingly parallel roles in several host-microbe interactions, both pathogenic and mutualistic. MAMPs are relatively conserved among bacteria, and hosts ranging from plants to animals have evolved mechanisms for MAMP recognition. Although the signaling roles of LPS, PG, and bioluminescence were discovered separately, and they each have distinct influences on the host, their effects appear intertwined and difficult to deconvolute. This is well illustrated by their combined influence on the developmental program associated with regression of the ciliated fields.
It was first discovered that LPS from the bacteria triggers apoptosis in the ciliated field but not regression of this structure (Foster et al. 2000). Specifically, the lipid A moiety of LPS had this effect—an intriguing finding, given that lipid A triggers responses in other host-microbe systems. Lipid A from V. fischeri has several modifications including a novel acylated phosphoglycerol moiety (Phillips et al. 2011) which could contribute to specificity, and at least one of the lipid A modifying enzymes, designated HtrB1, appears to contribute to colonization efficiency early in infection (Adin et al. 2008a). Moreover, a mutant of V. fischeri defective for the response regulator GacA has an altered LPS structure and is impaired in stimulating apoptosis and regression of the ciliated appendages on the light organ (Whistler et al. 2007). Apoptosis in the ciliated fields of the light organ does not specifically require V. fischeri lipid A, as LPS and lipid A from other bacterial species will also elicit apoptosis in these cells; however, the structure of lipid A does appear to influence its bioactivity in such assays (Foster et al. 2000). It is tempting to speculate that a distinctive lipid A structure is recognized by a host receptor(s), but it should be kept in mind that in natural infections, alterations in LPS structure could also influence colonization levels and membrane integrity, thereby affecting how much lipid A is presented to the host. Thus, structural elements of lipid A could affect either direct interactions with a host receptor or the amount of lipid A presented to host receptors.
In reporting the effects of LPS on the ciliated fields, Foster et al. (2000) noted that there must be at least one other signal and suggested potential candidates. One of these, PG, proved to be a second critical MAMP. PG stimulates mucus shedding by the ciliated epithelial fields (Nyholm et al. 2002), and it dramatically affects morphogenesis and regression of the ciliated fields (Koropatnick et al. 2004). Interestingly, it was also discovered that V. fischeri sheds a particular PG monomer that is usually recycled and kept within cells (Koropatnick et al. 2004). In two other Gram-negative bacteria, Bordetella pertussis and Neisseria gonorrhoeae, the same molecule is released from cells, and in each case, this molecule affects ciliated host cells. Indeed, this PG monomer is called tracheal cytotoxin (TCT), because it triggers the death of ciliated host airway cells in Bordetella infections, giving rise to its “whooping cough” symptoms. Although V. fischeri appears capable of TCT recycling, the combined activity of lytic transglycosylases apparently results in relatively large amounts of free TCT being released from cells (Adin et al. 2009). Using a mutant with decreased TCT release, Adin et al. (2009) provided evidence that the advantage of TCT shedding for V. fischeri may be that by triggering regression of the infection-promoting ciliated fields, an infecting strain thereby minimizes the chances of competition from later infecting V. fischeri strains.
Developmental changes appear to require the combined effect of the TCT and LPS signals. Koropatnick et al. (2004) found that TCT could stimulate hemocyte trafficking into the sinuses of the ciliated appendages as well as their regression, but curiously did not cause apoptosis in these cells. When TCT or PG was combined with LPS, however, the two had a synergistic effect on apoptosis and regression of the ciliated appendages that was very similar to that of a natural symbiotic infection (Koropatnick et al. 2004). Similarly, TCT and LPS together, but not singly, led to a decrease in NOS activity and NO similar to that seen in animals infected with V. fischeri (Altura et al. 2011). Interestingly, experiments with NOS inhibitors and NO donors suggested that NO itself may be an additional key signaling molecule in the apoptosis and morphogenesis associated with TCT and LPS exposure (Altura et al. 2011). Such two- or three-part signals may contribute to the specific recognition of the correct symbiont species.
Somewhat in contrast to these synergistic effects of LPS and TCT, one recent study suggested that TCT alone can in fact trigger apoptosis in the absence of LPS (Troll et al. 2009a). One explanation for this apparent discrepancy may be that assays measuring different stages of apoptosis were used in these two studies. TCT alone may stimulate initial elements of the apoptotic developmental program, but the combined effect of TCT and LPS likely represents the genuine symbiotic signal in a natural infection.
The recognition of MAMPs, including PG and LPS, by various hosts has been the subject of many studies, and homologs of known MAMP receptors and MAMP-responsive proteins have been found in E. scolopes. These MAMP-associated host genes include components of the NF-κB pathway (Goodson et al. 2005), specific PG recognition proteins (PGRPs) (Troll et al. 2009a, b), and LPS-binding proteins (Krasity et al. 2011). Although these MAMP-associated genes and proteins are orthologs of those in other organisms, they appear to have novel functions in E. scolopes. Notably, EsPGRP1 has an unprecedented nuclear localization, and infection by V. fischeri or treatment with TCT triggers loss of EsPGRP1 from host nuclei (Troll et al. 2009a). In contrast, EsPGRP2 is exported from host cells in association with light-organ mucus shedding; although similar to the dynamics of EsPGRP1, the export of EsPGRP2 is stimulated by TCT or infection with V. fischeri (Troll et al. 2009b). EsPGRP2 has an amidase activity capable of degrading TCT, and it is exported into the light-organ crypts colonized by V. fischeri, leading to the suggestion that it may attenuate this potentially toxic bacterial MAMP signal after it is received (Troll et al. 2009b).
The combined effects of TCT and LPS mimic many aspects of natural V. fischeri infection but as Troll et al. noted (2009a), they fail to completely duplicate it. An intriguing third signal may be the bioluminescence produced by V. fischeri. The squid appear capable of perceiving bioluminescence in the light organ, and components of photoreceptor-mediated signaling are present in light-organ tissue (Tong et al. 2009). Moreover, the host transcriptional profile varies depending on whether or not V. fischeri symbionts are bioluminescent (Chun et al. 2008). Although it is difficult to know to what extent these transcriptional changes are due to the perception of light itself or some other physiological change in dark symbionts related to the lack of luciferase activity, the presence of photoreceptors and the importance for the animal to match the intensity of its light-organ luminescence to the environment make a compelling case for light itself being perceived by the host. In any case, regression of the ciliated fields, export of EsPGRP2, and trafficking of hemocytes to the ciliated appendages early in infection are all attenuated relative to wild-type infections when squid are infected by dark mutants (McFall-Ngai et al. 2011). These differences are apparent even during infection initiation when the dark mutant presumably is colonizing at similar levels as the parent (Visick et al. 2000). Interestingly, among the host genes differentially regulated in wild-type and dark mutant infections are the PG recognition protein EsPGRP1 and a putative LPS-binding protein (LBP1) (McFall-Ngai et al. 2011). Light production induces transcription of the genes for these predicted MAMP receptors, as demonstrated by four- to fivefold lower levels of these transcripts in a (luxA) luminescence mutant (Chun et al. 2008). These data suggest that light production signals the host to boost production of receptors for PG and LPS.
Some of the developmental events occurring within the light organ and triggered by V. fischeri are not as well understood as those described above. In particular, changes in the morphology of epithelial cells lining the crypts and ducts, as well as proliferation of microvilli, have not been studied to the same extent as the developmental program associated with regression of the ciliated fields. This research focus probably reflects the relative difficulty in assaying changes in crypt and duct structure, which experimentally usually involves fixation, sectioning, and observation by TEM. It is known that crypt epithelial cell swelling requires V. fischeri to be bioluminescent (Visick et al. 2000), but microvillar proliferation, which is another process that can be reversed by curing symbionts, is unaffected by luminescence (Lamarcq and McFall-Ngai 1998). The swelling of epithelial cells resembles a response to hypoxic stress and could be tied to ongoing oxygen consumption by bioluminescence. Cell swelling could also represent a developmental response of the animal to light itself or to metabolic products related to the physiology of bioluminescence. Similarly, the proliferation of microvilli may relate to metabolic exchange. Although this is purely speculative, it would be consistent with the requirement for metabolically active symbionts to trigger and maintain an effect.
Bacterial Genes and Phenotypes Involved in Colonization
Bioluminescence and Pheromone-Dependent Regulation
As noted above, bioluminescence is the central contribution V. fischeri makes to this symbiosis (Fig. 20.7a , b ), and it appears intimately involved in the host response to the bacteria as well. V. fischeri has long been a model for studying bacterial bioluminescence, and the biochemistry, genetics, and regulation of light production, as well as the symbiotic role of bioluminescence, have all been active research topics. These areas are interrelated, and together their study has had a widespread influence on our understanding of bacterial gene regulation and host-bacterium interactions. For example, luminescence demands a large energetic commitment, which explains why the process is tightly regulated, and control of luminescence is accomplished in part by a pheromone-mediated regulatory pathway, which has become an archetype for understanding similar regulatory mechanisms in numerous host-associated bacteria.
Bioluminescence control in V. fischeri. (a and b) Images of the same juvenile light organ taken with light and in the dark, respectively; the light emitted by the bacteria within the organ can be seen in the center of the image taken in the dark. (c) The substrates and products in the reaction carried out by luciferase (LuxAB) to produce light. (d) The lux operon and the control of lux transcription by AI-modified LuxR. (e) The structures of the three pheromones produced in V. fischeri by the three pheromone synthases LuxI, AinS, and LuxS. Panels a and b have been published previously (Stabb 2005)
V. fischeri produces light when luciferase (LuxA and LuxB) converts FMNH2, in the presence of O2, and an aliphatic aldehyde (RCHO) to FMN, releasing water and an aliphatic acid (Fig. 20.7c ) (Hastings and Nealson 1977; Tu and Mager 1995; Ziegler and Baldwin 1981). LuxD produces RCHO, which is also generated by LuxC and LuxE through recycling of the corresponding aliphatic acid (Boylan et al. 1989). LuxG and other enzymes re-reduce FMN (Zenno and Saigo 1994; Nijvipakul et al. 2008). The obvious energetic costs of this process have stimulated curiosity in its regulation and usefulness for the bacteria. These costs include Lux protein biosynthesis [LuxAB can comprise 5% of the protein in bright cells (Hastings et al. 1965)], ATP hydrolysis during RCHO recycling, and the consumption of oxygen and reducing equivalents, which theoretically competes with energy recovery from aerobic respiration.
Primarily for energetic reasons, it has long been postulated that bioluminescence should slow the growth of V. fischeri. Consistent with this idea, some researchers found luminescence to be negatively correlated with growth in culture (Keynan and Hastings 1961; Hastings and Nealson 1977; Dunlap et al. 1995; Pooley et al. 2004), although the undefined nature of the luminescence mutants in these experiments raised some doubts as to whether luminescence was the only variable. Recently, however, it was shown that defined mutants lacking luxCDABEG could outgrow an isogenic parent under certain culture conditions, such as in a carbon-limited chemostat (Bose et al. 2008).
In contrast, bioluminescence is beneficial for V. fischeri colonizing the E. scolopes light organ. To begin with, the advantage of luminescence for the host (e.g., antipredatory camouflage) would confer a fitness advantage on the bacteria, because as noted above, V. fischeri populations appear augmented by successful symbiosis with the squid. However, the advantage for V. fischeri of expressing bioluminescence in the symbiosis goes deeper, as luminescence actually promotes successful colonization (Visick et al. 2000; Bose et al. 2008). Mutants lacking bioluminescence are able to colonize the host, but do not persist as well as their parental strains.
Several explanations have been proposed for how bioluminescence might aid bacteria directly (Stabb 2005). For example, it was proposed that bioluminescence stimulates photolyase-mediated DNA repair (Czyz et al. 2003); however, photolyase mutants are unimpaired in colonizing E. scolopes (Walker et al. 2006). As noted above, the host can apparently detect and respond to luminescence in its light organ (Chun et al. 2008; Tong et al. 2009), and it has been suggested that the squid might impose sanctions on dark infections, thereby ensuring that it receives bioluminescence in exchange for the nutrients it provides to V. fischeri. Other hypotheses have posited that the real function of luminescence is either to burn oxygen or to consume excess reductant (Bourgois et al. 2001; Visick et al. 2000). The latter idea now appears inconsistent with the regulation of luminescence (Bose et al. 2007; Lyell et al. 2010), but the model that luminescence confers an advantage by consuming oxygen is intriguing: by consuming oxygen, V. fischeri might generate hypoxic stress in the nearby host epithelium, attenuate ROS generation by the host, or render itself more resistant to ROS. Luciferase’s high affinity for O2 (Km = ~35 nM) is consistent with a role in driving down ambient or intracellular oxygen levels (Bourgois et al. 2001). The ideas that the host sanctions dark infections or that the light-organ environment somehow makes bioluminescence physiologically beneficial are also both plausible and are not mutually exclusive.
The lux genes underlying light production are regulated tightly and induced in the symbiosis. Key enzymes and regulators for light production in V. fischeri are encoded in an 8-kb DNA locus (Engebrecht et al. 1983; Engebrecht and Silverman 1984) including the luxICDABEG operon and the divergently transcribed luxR gene (Fig. 20.7d ). luxCDABEG encodes the enzymes for luminescence, while luxI encodes a pheromone synthase and luxR encodes a corresponding pheromone receptor. Specifically, LuxI generates N-3-oxo-hexanoyl homoserine lactone (3-oxo-C6-HSL) (Fig. 20.7e ) (Schaefer et al. 1996), a pheromone that diffuses across the cell membrane (Kaplan and Greenberg 1985). As cell density increases, 3-oxo-C6-HSL accumulates until it reaches a critical concentration where it can bind LuxR, producing an activator that binds a “lux box” sequence to activate transcription of luxICDABEG through interactions with RNA polymerase that compensate for a poor -35 promoter element (Egland and Greenberg 1999; Fuqua et al. 1994). The result is that the lux genes are most highly expressed at high cell densities, when the bacteria have reached a “quorum” (Fuqua et al. 1996). Moreover, like many bacterial pheromone-mediated regulatory systems, the lux operon constitutes a positive feedback circuit, because the 3-oxo-C6-HSL produced by LuxI leads to increased transcription of luxICDABEG. Consequently, environmental regulatory inputs to the lux system can be amplified, and in theory, the response could spread through a population.
It has now become clear that pheromone-mediated regulation of bioluminescence in V. fischeri is quite a bit more complex than this canonical model. Two other pheromones also modulate the lux genes (Fig. 20.7e ); N-octanoyl homoserine lactone (C8-HSL) produced by AinS (Hanzelka et al. 1999; Kuo et al. 1994, 1996) and “AI-2,” which is produced by LuxS (Lupp and Ruby 2004) and is presumably a furanosyl borate diester as it is in Vibrio harveyi (Chen et al. 2002). AI-2 and C8-HSL apparently function through distinct receptors that both act via LuxU and LuxO, Hfq, and a small RNA called Qrr to increase levels of LitR, which in turn increases luxR expression (Fidopiastis et al. 2002; Lupp et al. 2003; Miyamoto et al. 2000; Miyashiro et al. 2010). C8-HSL can also bind to and activate LuxR, although it is less effective than 3-oxo-C6-HSL and can even inhibit 3-oxo-C6-HSL-mediated stimulation (Egland and Greenberg 2000; Schaefer et al. 1996). As with LuxI and LuxR, there is also positive feedback in the AinS-AinR system (Lupp and Ruby 2004). These interconnected regulatory systems were reviewed recently elsewhere (Stabb et al. 2008).
3-oxo-C6-HSL, C8-HSL, and AI-2 all increase in concentration with increasing cell density, and textbook models of pheromone-mediated regulation typically depict these pheromones as a mechanism of cell-density-dependent regulation called quorum sensing. However, in V. fischeri and many other systems, it is clear that pheromone-mediated signaling is not simply a census-taking process (Dunn and Stabb 2007). Both the pheromone synthases and the pheromone receptors are also regulated in response to environmental conditions, such that high cell density may be necessary to elicit a behavior, but a “quorum” is typically not sufficient for a full response unless other conditions are met.
Context dependence of pheromone-mediated signaling is obvious in V. fischeri strains isolated from the light organs of E. scolopes. Such strains induce pheromone synthesis and luminescence in the host light organ, but they are dim and produce less pheromone in culture (Boettcher and Ruby 1990; Lee and Ruby 1994a), even at equivalent cell densities (Stabb, unpublished data). Such context-dependent lux expression was evident even in comparisons of different light-organ microenvironments: when V. fischeri was marked with both a constitutive red fluorescent protein gene and a luxI promoter-gfp reporter, dense populations of bacteria appeared in the three distinct light-organ crypt types in a similar time frame, but lux induction lagged in crypt 3 (Dunn et al. 2006), which is also the last crypt to mature developmentally (Montgomery and McFall-Ngai 1993). These data are consistent with induction of luminescence by a host-derived environmental cue initially absent in crypt 3. Clearly, high cell density may be necessary for pheromone-mediated induction of luminescence in V. fischeri, but it is not sufficient.
Bioluminescence and pheromone production by V. fischeri respond to the environment; however, the underlying regulatory mechanisms are still being elucidated. This is particularly true for V. fischeri strain ES114, which serves as a model strain typical of other E. scolopes symbionts (Boettcher and Ruby 1990; Lee and Ruby 1994a). Most early studies of lux regulation used either bright V. fischeri strains such as MJ1, which was isolated from a pinecone fish (Ruby and Nealson 1976), or the lux region cloned in Escherichia coli. The core circuitry of lux in ES114 is similar to that of MJ1 (Gray and Greenberg 1992b; Gray and Greenberg 1992a), but like most isolates from E. scolopes, ES114 is dim in culture.
With the caveat that there could be strain-specific differences, environmental control of lux in V. fischeri has been studied with respect to different regulators. For example, cAMP receptor protein (CRP)-mediated activation of lux was documented using transgenic lux-containing E. coli (Dunlap and Greenberg 1985, 1988; Dunlap and Ray 1989), and it was similarly shown that V. fischeri strain MJ1 regulates luminescence in response to glucose, a phenomenon that may be CRP-mediated (Friedrich and Greenberg 1983). Luminescence of MJ1 is also inhibited by iron (Haygood and Nealson 1985).
Recently, the redox-responsive ArcA/ArcB two-component regulatory system was identified as a strong repressor of luminescence (Bose et al. 2007; Lyell et al. 2010). Both arcA and arcB mutants are ~500-fold brighter in culture than ES114, and activated ArcA binds the luxI promoter near the “lux box” (Bose et al. 2007). In E. coli, the Arc system is activated by reducing conditions, and consistent with this mechanism, luminescence is induced by relatively oxidizing conditions (Bose et al. 2007; Stabb et al. 2008). Interestingly, however, ArcA/ArcB does not significantly repress luminescence during symbiotic colonization, and arcA mutants achieve nearly the same brightness in culture as wild-type cells do in the host. Thus, regulation by ArcA/ArcB could account for most of the differences in luminescence observed between cultured and symbiotic cells by invoking a model in which Arc represses lux in culture, but luminescence is derepressed by inactivation of Arc during initial infection (Bose et al. 2007). On the other hand, the ArcA/ArcB system could be active and repressing lux during initiation of the symbiosis but simply overpowered by another regulatory effect.
Other genes affecting luminescence in V. fischeri have been reported (Hussa et al. 2007; Visick et al. 2007; Whistler and Ruby 2003). Recently, a mutant screen revealed numerous loci involved in regulation of the luxICDABEG operon, either directly or indirectly (Lyell et al. 2010). That study also revealed environmental conditions that influence luminescence of wild-type V. fischeri. For example, mutants lacking a phosphate-uptake system or phoQ were brighter than wild type, leading to the demonstration that phosphate or Mg2+ levels can influence luminescence of wild-type cells (Lyell et al. 2010). Remaining challenges include determining which environmental conditions that influence luminescence in culture also impact symbiotic luminescence and whether information about environmental conditions can be communicated among V. fischeri cells using pheromone signaling, as the results thus far would seem to indicate.
It should also be noted that luminescence is just one part of the pheromone-controlled regulon in V. fischeri, and the role(s) of other genes co-regulated by C8-HSL, 3-oxo-C-HSL, and AI-2 along with luminescence should be enlightening. LuxR controls several genes besides the luxICDABEG operon (Antunes et al. 2007, 2008; Callahan and Dunlap 2000), and some of the corresponding proteins were identified in the proteome of symbiotic V. fischeri (Schleicher and Nyholm 2011). Especially noteworthy is the novel protein QsrP, which has no known function but is induced by LuxR and was among the most abundant proteins in symbionts (Schleicher and Nyholm 2011). Visick et al. (2000) demonstrated that the colonization deficiency of luxR mutants could be overcome by expressing luxCDABEG independently of this activator, suggesting that bioluminescence is the major (or only) LuxR-dependent colonization factor; however, other LuxR-regulated genes could have subtle influences on the symbiosis or major effects on symbiotic colonization but subtle degrees of LuxR-mediated regulation. Also, C8-HSL and AI-2 influence the expression of more genes than luxR, and the AinS/AinR regulon in particular is distinct from that of LuxR and important in host colonization (Lupp and Ruby 2004, 2005). Understanding the interplay of these three pheromone systems in the light-organ symbiosis, their control by the symbiotic environment, the dynamics of their signaling, and the constituents of their respective regulons should be a rich area for further research.
Biofilm Formation and Biofilm Regulators
Studies of the initiation of symbiotic colonization revealed that an early stage involves the attachment of V. fischeri to the surface of the light organ in a biofilm-like aggregate (Fig. 20.2d ). Subsequent studies began to probe the mechanism of that attachment. In particular, a search for genes important in the symbiosis revealed a requirement for rscS (regulatory of symbiotic colonization, sensor) (Visick and Skoufos 2001), which encodes a hybrid sensor kinase protein, and syp (Yip et al. 2005), an 18-gene symbiosis polysaccharide locus. It was subsequently determined that these genes are necessary both for the production of biofilms in culture and for the formation of the symbiotic aggregate (Yip et al. 2006) (Fig. 20.8 ).
Control over biofilm formation by the syp regulators. The activities of two sensor kinases, RscS and SypF, influence syp-dependent biofilm formation. Sensor kinases respond to an environmental signal by autophosphorylating and donating a phosphoryl group to a downstream response regulator. In the case of the syp locus, phosphorylated SypG is predicted to activate transcription from four promoters, resulting in the production of a Syp-produced polysaccharide that promotes biofilm formation. Biofilm formation can be visualized in the laboratory by the development of wrinkled colonies and the production of pellicles at the air/liquid interface of static cultures (which can be strong enough to retain liquid when the test tube is inverted), and in symbiosis by the production of large aggregates on the surface of the light organ. RscS also appears to inactivate SypE, a response regulator that exerts a minor positive and a strong negative impact on biofilm formation. The role of SypF is poorly understood, but it appears to act upstream of SypE/SypG and VpsR to impact syp-dependent biofilm formation and cellulose biosynthesis, respectively (This model, which was previously published (Visick 2009), has been slightly altered by the addition of an arrow extending from SypE to biofilm formation, indicating the slight positive effect exerted by SypE on biofilm formation (Morris et al. 2011))
Mutants lacking RscS are defective in initiating symbiotic colonization: most animals inoculated with this mutant remain uncolonized, while others exhibit a delay in initiating colonization (Visick and Skoufos 2001). This defect can be attributed to the inability of this mutant to aggregate on the surface of the light organ (Yip et al. 2006). rscS encodes a sensor kinase protein, a fact that immediately suggested a model for its function in symbiosis. Sensor kinases work by sensing an environmental signal, autophosphorylating on a conserved histidine residue, and serving as a phospho-donor to a downstream response regulator. The activated response regulator then carries out a response, most commonly acting as a transcription factor to increase or decrease transcription of genes. Although in many cases the genes for a sensor kinase/response regulator pair are physically linked in the chromosome, this was not the case for rscS. Subsequent studies revealed that RscS acts upstream of the response regulator SypG to control syp transcription and biofilm formation (Hussa et al. 2008).
Mutants defective for syp also exhibit defects in initiating symbiotic colonization. The syp locus comprises 18 genes (sypA-R), four of which encode regulatory proteins, while the remaining genes in the cluster encode proteins with predicted functions in polysaccharide biosynthesis, modification, and export (Yip et al. 2005). The genes in the syp locus are grouped into at least four operons (Fig. 20.8 ), each of which contains promoter sequences for transcription by RNA polymerase carrying the alternative sigma factor σ54. Transcription from such promoters depends upon binding and activation by a σ54-dependent transcriptional activator, which typically binds upstream of the promoter and provides the energy for open complex formation (Buck et al. 2000). Indeed, a conserved sequence located upstream of each of the syp promoters has been identified and may serve as the binding site for this key activator. The response regulator SypG is likely to serve as the activator for transcription of the syp locus, as this protein contains a putative σ54 interaction domain in addition to its N-terminal receiver (REC) domain (containing the site of phosphorylation) and C-terminal DNA-binding domain. In support of this idea, overexpression of SypG promotes syp transcription in a manner that depends upon the rpoN gene, which encodes σ54 (Yip et al. 2005).
The similarity in the symbiotic phenotypes of rscS and syp mutants led to the hypothesis that these proteins work in the same pathway, specifically, that RscS controls transcription of the syp locus. Indeed, overexpression of RscS led to an increase in syp transcription, and this induction depended on SypG (Fig. 20.8 ) (Hussa et al. 2008; Yip et al. 2006). Excitingly, overexpression of RscS also led to a number of unusual biofilm phenotypes (Yip et al. 2006). Whereas wild-type cells form smooth colonies on complex solid media, RscS-overexpressing cells form colonies with a wrinkled morphology (i.e., with 3D architecture), consistent with biofilm formation. In addition, RscS-overexpressing cells form a biofilm pellicle at the air-liquid interface during static growth in a minimal medium, while control cells do not. These phenotypes also depend upon SypG. Finally, consistent with the idea that RscS and Syp contribute to colonization through their role in symbiotic aggregate formation, overexpression of RscS induces the formation of very large aggregates on the light-organ surface (Fig. 20.8 ). Increased aggregate formation depended upon an intact syp locus: mutation of a representative syp gene, sypN, which encodes a putative glycosyltransferase predicted to be involved in the production of a polysaccharide, disrupted aggregate formation (Yip et al. 2006). The formation of this large aggregate is not detrimental to colonization; rather, RscS-overexpressing cells fully outcompete control cells containing an empty vector for symbiotic colonization.
The syp locus is controlled by several regulators besides RscS and SypG. Currently, the best understood of these additional regulators is SypE, a second response regulator encoded within the syp locus. SypE is an unusual response regulator in that its REC domain, containing the putative site of phosphorylation, is centrally located, rather than localized to the N-terminus as is typical of other response regulators. The REC domain is flanked by putative serine kinase (N-terminus) and serine phosphatase (C-terminus) domains (Morris and Visick 2010). SypE appears to act as both an inhibitor and an activator of biofilm formation, depending on the conditions used to promote biofilm formation (overexpression of RscS or SypG) (Hussa et al. 2008). Such phenotypes are consistent with opposing activities for the terminal domains. Indeed, it is now known that the putative serine kinase domain inhibits biofilm formation, while the putative serine phosphatase activates it (Morris et al. 2011).
These data led to a model in which RscS, in addition to activating SypG to promote syp transcription, inactivates SypE to permit biofilm formation. Given the nature of these proteins, Morris et al. (2011) predicted that RscS promotes phosphorylation of SypE, thereby inactivating it. Consistent with that prediction, a substitution of alanine for aspartate at the predicted site of phosphorylation results in a protein with constitutive inhibitory activity, presumably because it is unable to be phosphorylated (Morris et al. 2011). Interestingly, a sypE mutant does not exhibit a colonization defect, perhaps due to SypE’s subtle positive role in biofilm formation. However, expression of the SypE mutant protein that cannot be phosphorylated and thus inactivated causes a severe defect in symbiotic colonization. Therefore, it appears that SypE inhibits symbiotic colonization and must be inactivated to permit colonization, presumably by RscS. The symbiotic defect of an rscS mutant could be attributed, in part, to its failure to inactivate SypE. Consistent with this model, an rscS sypE mutant colonized better than an rscS mutant. Thus, productive symbiosis depends upon inactivation of a negative regulator.
Another regulator encoded by the syp locus is SypF. Neither insertional disruption of sypF nor overexpression of wild-type SypF impacted biofilm formation (Darnell et al. 2008). However, overexpression of an allele with increased activity, sypF*, led to increased biofilm formation, including wrinkled colony formation and pellicle production. Surprisingly, these phenotypes persisted even when the two syp response regulators, sypG and sypE, were mutated. However, biofilm formation was eliminated when both sypG and a second response regulator, vpsR, were disrupted (Fig. 20.8 ). In V. cholerae, VpsR controls the vps polysaccharide locus, which is only poorly conserved in V. fischeri (Yildiz et al. 2001; Yildiz and Visick 2009; Darnell et al. 2008). Instead, it appears that a locus involved in cellulose biosynthesis contributes to biofilm formation induced by overexpression of SypF*. Clearly, control of biofilm formation in V. fischeri is complex and is not limited to a single regulator.
Many questions remain unanswered. How SypF is integrated into the regulatory pathway remains unclear, and its role in symbiotic colonization has not yet been established. The product of the syp locus is thought to be a polysaccharide (Yip et al. 2006), but the composition of the polysaccharide is as yet undetermined. Importantly, the environmental signals that activate RscS and the other regulators to induce biofilm formation and initiate symbiotic colonization are as yet unknown. Moreover, since symbiotic colonization is not impaired by overproduction of RscS and enhanced aggregate formation, there must be a yet-undiscovered mechanism by which aggregation is turned off to permit dispersal and migration into the pores for colonization to proceed. The discovery of the syp locus and its role in biofilm formation and initiating symbiotic colonization has provided important insights into host colonization by V. fischeri and an important model for in vivo biofilm formation and will continue to raise new questions to direct future research.
Motility, Chemotaxis, and Their Regulation
V. fischeri is motile by virtue of a polar tuft of flagella (Fig. 20.9a ) (Ruby and Asato 1993). Early on in the study of this symbiosis, it was determined that motility of V. fischeri is essential for symbiotic colonization; however, the roles of motility, flagellation, and chemotaxis continue to be intriguing and outstanding questions. Initial studies performed with uncharacterized mutants revealed that both nonflagellated and flagellated-but-nonmotile (paralyzed) mutants fail to colonize (Graf et al. 1994). However, motility per se is not required for the aggregation stage (Nyholm et al. 2000; Millikan and Ruby 2002), and once inside the crypt spaces, most V. fischeri cells become nonflagellated (Fig. 20.9b ) (Ruby and Asato 1993). It is not known whether the loss of flagella is critical for colonization, especially since some bacteria do retain motility (Millikan and Ruby 2003). However, it is likely that flagella could interfere with the ability of the cells to achieve high cell density in the crypt spaces. Finally, V. fischeri cells rapidly regain their flagella upon release into seawater (Fig. 20.9b ) (Ruby and Asato 1993).
Role of motility in symbiosis. (a) V. fischeri contains a tuft of polar flagella. (b) Presence of flagella during different stages of colonization. Nonmotile cells are competent to aggregate, but motility is required for entry into the light-organ pores and presumably for passage to the crypts. Upon colonization, many bacteria lack flagella. Finally, following release from the light organ, V. fischeri cells can rapidly regrow their flagella. (c) Model for regulatory control by FlrA, a σ54-dependent regulator. FlrA is predicted to induce transcription of the genes for the two-component system FlrBC, which, along with σ54, controls transcription of the gene for the major flagellin protein FlaA. FlrA is also predicted to control transcription of the gene for the alternative sigma factor, FliA, which likely directs transcription of the genes for the other flagellin proteins, FlaB-E and potentially FlaF. Finally, FlrA appears to positively and negatively control the production of other proteins, although whether this effect is direct is unknown. The figure in panel A was previously published (O’Shea et al. 2005), and the scale bar represents 200 μM (The regulatory scheme depicted in panel C is modeled from reported data for V. fischeri and V. cholerae (Millikan and Ruby 2003; Prouty et al. 2001))
One study suggested that excessive amounts of flagella are detrimental to initiation of colonization. Millikan and Ruby isolated three classes of hypermotile mutants with increased motility in both liquid and semisolid media (Millikan and Ruby 2002). The increased motility apparently stemmed from an increase in the number of flagella, from 1 to 3 on wild-type cells to upwards of 16 flagella on the mutants. Mutants in classes II and III also produced mucoid colonies rather than the smooth nonmucoid colony of their parent furthermore; class II mutants had a growth defect, and class III mutants exhibited defects in bioluminescence, 3-oxo-C6-HSL pheromone production, and the ability to hemagglutinate red blood cells. All three classes exhibited defects in initiating symbiotic colonization, failing to reach wild-type levels in 24 h. Class III mutants demonstrated the most severe defect, achieving only 0.1–10% of wild-type levels at 24 and 48 h postinoculation. The defect in initiation was attributed to deficiencies at the aggregation stage: wild-type cells form non-motile aggregates within 4–6 h; in contrast, the mutants retained motility. Furthermore, aggregate formation by the mutants was delayed, occurring between 8 and 12 h after inoculation, and aggregates were significantly smaller (tens of cells rather than hundreds) than those formed by the wild type (Millikan and Ruby 2002). Although the mutations in these strains were never identified, these studies nevertheless indicate the importance of regulating motility during entry into the symbiosis and beyond.
Subsequent studies and the availability of the genome sequence have confirmed the importance of motility in this symbiosis and have identified specific pathways of genetic control. Almost all genes predicted to be involved in flagellar assembly and regulation are housed together in a single locus of about 60 genes on the larger of V. fischeri’s two chromosomes. The functions of these genes can be readily predicted through comparisons with well-studied models such as E. coli and Salmonella (Chevance and Hughes 2008). Furthermore, comparisons with other organisms suggest that regulation of flagellar genes in V. fischeri is likely hierarchical. Perhaps the best comparison is with the closely related microbe V. cholerae, which has been extensively studied (Correa et al. 2000, 2005; Prouty et al. 2001). In that organism, the master flagellar regulator, FlrA, controls a number of flagellar genes, including additional regulators such as FlrBC and FliA. FlrBC is a predicted two-component signal transduction system that controls the next level of flagellar genes, including flaA, a flagellin gene; flagellin protein constitutes the major subunit assembled into the external filament of the flagellum. Indeed, a V. fischeri flrC mutant is non-motile and, like other non-motile mutants, exhibits a colonization defect (Hussa et al. 2007). FliA, or σ28, activates another subset of flagellar genes, including four other flagellin genes (flaB-E). The working model is that V. fischeri likely controls its flagella using a hierarchy similar to that of V. cholerae (Fig. 20.9c ) (Millikan and Ruby 2003; Prouty et al. 2001).
In V. fischeri, the role of FlrA was investigated in detail by Millikan and Ruby (2003). flrA mutants are non-motile, as predicted from FlrA’s putative role as master flagellar regulator, and fail to colonize. Surprisingly, complementation with FlrA expressed from a plasmid restored normal motility in culture but not normal colonization: complemented flrA mutants failed to initiate colonization at the rate and with the success of wild-type cells (only 35–46% of animals became colonized by the complemented flrA mutant relative to 94% of wild-type-inoculated animals); they also failed to achieve wild-type levels of colonization (1.6 × 103 colony-forming units of flrA mutant cells per squid relative to 9 × 104 colony-forming units of wild-type bacteria per squid). These experiments suggest that the proper level or location of FlrA is critical for symbiosis, and indicate that a trait other than motility, which was restored by multi-copy expression of flrA based on in vitro assays, may be responsible for the symbiotic defect in the flrA mutants (Millikan and Ruby 2003). In support of this idea, symbiotic aggregates produced by flrA mutants are diminished in size relative to those produced by the wild-type strain (tens of cells vs. hundreds of cells), despite the fact that other non-motile mutants aggregate normally (Nyholm et al. 2000). Millikan and Ruby subsequently identified four nonflagellar genes that were controlled by FlrA positively (e.g., genes encoding topoisomerase and halovibrin C) or negatively (e.g., genes encoding phosphoglycerate kinase and potassium efflux protein) (Fig. 20.9c ). Whether FlrA controls these genes directly, and whether these FlrA-regulated genes play roles in symbiosis, remains unknown.
The FlrA protein contains a σ54 interaction domain and a DNA-binding domain and thus is predicted to activate transcription of target genes in conjunction with σ54. Indeed, rpoN mutants, which lack σ54, are non-motile, fail to induce transcription of flaA, and fail to initiate colonization (Millikan and Ruby 2004; Wolfe et al. 2004). Like FlrA, σ54 controls additional genes involved in colonization, notably luminescence and the syp polysaccharide locus (Wolfe et al. 2004; Yip et al. 2005). The use of this alternative sigma factor is of note, as genes under the control of σ54 tend to be under tighter “on/off” regulatory control (Buck et al. 2000); whether or not this regulation is key for colonization remains to be determined.
Of V. fischeri’s six flagellin genes (flaA-F), only flaA appears to be directly regulated by FlrA and σ54. Despite the redundancy of flagellin genes, flaA mutants show reduced motility with fewer motile cells, and fewer flagella per cell. In contrast, a flaC mutant was seemingly unaffected for motility and flagellar elaboration. The flaA mutant also exhibited a symbiosis defect: loss of flaA delayed symbiotic colonization by about 3 h, and prevented full colonization (20–25% of that achieved by the wild-type strain) (Millikan and Ruby 2004). The mutant was also defective in competing for colonization with the wild type, and furthermore, it appeared to be preferentially expelled from the light organ. GFP-labeled cells revealed that, in contrast to the flrA regulatory mutant, the flaA flagellin mutant formed aggregates with the same approximate size as wild-type aggregates; furthermore, the flaA mutant entered the light organ normally. However, while the wild-type cells reached and colonized deep crypt spaces within 16 h postinoculation, flaA mutant cells did not colonize these sites within 20 h. This inability to reach and colonize the deep crypt spaces could account for both the delay in colonization and the preferential expulsion of the flaA mutant.
As suggested by the phenotype of the flaA mutant and numerous studies in other organisms, one prime reason for cells to be motile is to permit them to move to optimal locations. Typically, this movement is directed toward nutrients and attractants and away from toxic molecules and repellents using chemotaxis. The role of chemotaxis in symbiotic colonization by V. fischeri is currently an active area of investigation. One study asked what molecules serve as chemoattractants for V. fischeri (DeLoney-Marino et al. 2003). In tryptone-based soft agar media, V. fischeri forms two rings as it migrates from the spot of inoculation, indicating that it senses and responds to two different molecules that it likely consumes. The inner ring is comprised of cells sensing the amino acid serine. Cells in the outer ring sense the nucleic acid thymidine, which is present in tryptone. In addition to thymidine, V. fischeri can sense and respond to other ribonucleosides (adenosine, guanosine, uridine, and cytidine) as well as deoxynucleotide triphosphates. This ability of V. fischeri to sense nucleosides and nucleotides is unusual, although more recently it has been shown that E. coli and Pseudomonas putida can perform chemotaxis to pyrimidines (thymine and uracil in the case of E. coli, cytosine in the case of P. putida) (Liu and Parales 2008; Liu et al. 2009). In addition to serine and the building blocks of DNA and RNA, V. fischeri responds to a number of sugars, including N-acetylneuraminic acid, which is one of the sugars in the mucus secreted by the squid (DeLoney-Marino et al. 2003; Nyholm et al. 2000).
It remains unknown whether chemotaxis helps direct the bacteria into the light organ, and if so, which chemoattractants are used, but a chemotaxis-defective cheY mutant fails to compete effectively against wild-type cells for colonization (Hussa et al. 2007). In E. coli and Salmonella, CheY is a response regulator that modulates reversal of flagellar rotation, permitting “tumbles” that when interspersed with “runs” permit the cells to reorient and migrate toward nutrients and away from repellents. (Baker et al. 2006). Cells with defects in tumbling, such as cheY mutants, fail to perform normal chemotaxis; in addition, they can become trapped in “dead-end” passages present in agar-solidified media, making them appear nonmotile (Wolfe and Berg 1989). Like E. coli and Salmonella mutants, the V. fischeri cheY mutant exhibits smooth swimming, which may prevent it from appropriately migrating toward a stimulus. Alternatively, it may get stuck in squid-secreted mucus. Distinguishing between these possibilities requires the identification of a specific chemoreceptor(s) required for attractant recognition during symbiotic colonization. However, V. fischeri contains upwards of 40 such receptors, making this endeavor a challenge (Ruby et al. 2005).
One intriguing and outstanding question is: What factors direct the loss of flagella inside the light organ? Another is its corollary, What promotes flagellar regrowth in the nutrient-limited seawater? One possibility is suggested by the dependence of V. fischeri motility on magnesium (O’Shea et al. 2005). Cells grown in a medium lacking magnesium are poorly motile, as assessed on soft agar plates. Addition of either magnesium sulfate or magnesium chloride restores full migration. In particular, magnesium sulfate concentrations between 20 and 40 mM, similar to that found in seawater (about 50 mM), were optimal in promoting migration through soft agar. Other cations, such as calcium, could promote motility but to a lesser extent at low concentrations (5 mM); at concentrations higher than 10 mM, CaCl2 was inhibitory to growth and thus difficult to assess. Subsequent analysis revealed that cells grown without magnesium elaborated few to no flagella, while those grown with magnesium contained on average two to three flagella and as many as seven or eight (O’Shea et al. 2005). It is unknown how magnesium impacts flagellar production, although it does not appear to be at the level of transcription of flagellin genes (O’Shea et al. 2006).
A search for mutants that were motile in the absence of magnesium led to the identification of two genes encoding the diguanylate cyclases MifA and MifB. These proteins are involved in the production of cyclic diguanylate (c-di-GMP), which is known to influence motility (Wolfe and Visick 2010). Disruption of either mifA or mifB increased motility in the absence of magnesium. However, neither mutant alone nor a double mutant defective for both genes exhibited the same pattern of migration in the absence of magnesium as the wild-type strain exhibits in the presence of magnesium. Thus, it remains unclear how magnesium impacts motility.
Regardless of how magnesium acts to stimulate the production of flagella, seawater contains sufficient levels of magnesium to permit V. fischeri to express flagella and thus be competent to colonize its squid host. Once inside, it is possible that decreased levels of magnesium and/or calcium contribute to the mechanism that promotes loss of flagella on colonizing bacteria. The levels of magnesium and calcium inside the light organ of E. scolopes are presently unknown.
In summary, research to date has demonstrated the importance of flagella and motility to symbiosis, but the role of motility is not simple and straightforward. In other systems, flagella can serve as mediators of attachment, a possibility that is supported by the aberrant aggregation of the hypermotile and flrA mutant strains. In some hosts, flagellin proteins can act as MAMPs, in much the same way as PG and LPS, which is an intriguing possibility given the signaling obvious in this symbiosis. Flagellar components can also serve as type III secretion systems that direct bacterial proteins into the host environment, and this remains a possibility for V. fischeri as well. When motility or components of the flagellar system are required and when/if they are detrimental, when/if flagella or the flagellar apparatus contributes to attachment or secretion, what the precise contribution of chemotaxis and where the optimal locations of the bacteria are, and how flagella are lost and regained are all important questions that remain to be pursued. No doubt the answers will provide important insights into host colonization and colonization dynamics.
Iron Uptake
Eukaryotic hosts generally limit the availability of iron, which is necessary for the growth and metabolism of bacteria, as a defense mechanism (reviewed in (Nairz et al. 2010)). As a result, bacteria have evolved numerous systems for the acquisition of iron. V. fischeri, for example, encodes a number of different proteins predicted to be involved in binding of iron or iron complexes and their associated transport systems (Ruby et al. 2005). The first study examining the role of iron uptake during the V. fischeri-squid symbiosis suggested that, indeed, the ability to acquire iron was an important symbiosis trait (Graf and Ruby 2000). This report relied on a transposon mutagenesis screen for mutants with altered siderophore production and focused on one that appeared completely defective in its ability to sequester iron. Indeed, growth of this strain depended on the presence of iron in the medium. Surprisingly, this mutant turned out to contain an insertion not in a siderophore biosynthesis gene, but in glnD, a gene known in E. coli to play a role in sensing nitrogen status (reviewed in (Arcondeguy et al. 2001)). Consistent with a similar role for GlnD in V. fischeri, the glnD mutant was deficient in utilizing a variety of nitrogen sources. Complementation with a wild-type copy of glnD restored all of the observed phenotypes to wild-type levels, supporting the novel role of GlnD in siderophore production. Finally, the glnD mutant exhibited a defect in colonizing juvenile squid. In support of the idea that the symbiotic phenotype was due to the iron sequestration defect, the defect was more pronounced when cells in the starting inoculum were pre-grown under conditions of limiting iron availability. Furthermore, addition of iron to seawater alleviated the defect of the glnD mutant (Graf and Ruby 2000). The mechanism by which GlnD impacts siderophore production remains unknown.
A more directed study investigated the role of one possible mode of acquiring iron, heme transport, in the V. fischeri-squid symbiosis. Bioinformatic analyses identified a locus, VF_1220-VF_1228, similar to heme-uptake loci in other Vibrios (Septer et al. 2011; Wang et al. 2010a). One operon within this locus (VF_1225-VF_1220) is predicted to encode a periplasmic heme-binding protein (HutB), an inner membrane permease (HutC), an ABC-transporter ATPase (HutD), and proteins that function in providing the energy for transport (TonB, ExbB, ExbD). Additional proteins predicted to be involved in optimal heme utilization were encoded in a divergent operon (HutW, HutX, HutZ, VF_1226-VF_1228). These genes, as well as putative hemin receptor genes VF_1234 and VF_A0331-A0333, were downregulated upon exposure of wild-type cells to NO in a manner that depended upon an intact hnoX gene, which encodes an NO sensor (Wang et al. 2010a). Regulatory sequences consistent with control by Fur (Ferric uptake regulator) were identified.
Consistent with the putative role of this locus, its deletion disrupted the ability of V. fischeri to grow with hemin (Fe3+) as a source of iron, but did not impact growth of the cells when ferrous sulfate was supplied as an iron source (Septer et al. 2011; Wang et al. 2010a). Investigations into the transcriptional control of this locus revealed that two divergent promoters, upstream of VF_1225 and VF_1226, were upregulated in response to low iron availability. Furthermore, both promoters were repressed by Fur, as disruption of fur led to an increase in transcription even in the presence of iron. These data are consistent with a model in which low iron availability causes an increase in transcription of this heme-uptake locus; when iron levels rise, Fur is activated to decrease transcription of the locus (Septer et al. 2011). Thus, V. fischeri appears to carefully modulate transcription of this locus in response to iron availability. To assess the relevance of this phenomenon and these genes in symbiosis, two approaches were used. First, the transcriptional response of this locus in symbiosis was assessed with promoter-GFP reporters. Transcription of VF_1225 was turned on as early as 14 h postinoculation, indicating that the light-organ environment is low in iron. Second, the competence of the deletion mutant to colonize when presented in competition with the wild-type strain was evaluated. The mutant exhibited a defect in colonization, although the defect was not apparent until after several days postinoculation. Thus, these data support and extend the study by Graf and Ruby (2000) that the light-organ environment is limiting for iron, and indicate that uptake of heme is an important mechanism by which V. fischeri acquires iron during symbiosis. In further support of the importance of heme uptake in symbiosis, the transcriptome study by Weir et al. (2010) found that the heme-uptake genes were induced following bacterial venting. At venting, membrane “blebs” (presumably host derived) appear, and it was speculated that these contain a source of iron for the symbionts (Septer et al. 2011). Finally, a recent study investigating the proteomes of V. fischeri and E. scolopes identified host-iron-binding proteins, including transferrin, ferritin, and melanotransferrin, and symbiont heme-binding proteins (Schleicher and Nyholm 2011). Whether heme is the primary source of iron during symbiosis, and how/when iron is supplied to the bacteria, will be of interest to determine.
GacA
Another important regulatory protein involved in symbiosis is the response regulator GacA. A gacA mutant was defective in initiating symbiosis: only about 50% of animals became colonized under conditions in which 100% of wild-type-inoculated animals became colonized, and animals that became colonized with the gacA mutant contained on average 100X fewer bacteria (Whistler and Ruby 2003). Moreover, as mentioned above, in squid that were colonized by gacA mutants, the symbionts did not trigger some of the normal developmental events in the host that are elicited by wild-type V. fischeri, including apoptosis and cessation of mucus shedding in light-organ epithelial cells (Whistler et al. 2007). To take into account the possibility that low colonization by the gacA mutant might underlie these apparent non-effects on the host, comparisons were made to a lysA mutant, which is a lysine auxotroph that is also attenuated in colonization (Whistler et al. 2007). Using this control, the authors concluded that low colonization alone could not explain the altered signaling capacity of the gacA mutant.
In culture, V. fischeri GacA controls bioluminescence, motility, LPS structure, siderophore production, and nutrient acquisition (Whistler et al. 2007; Whistler and Ruby 2003). In rather broad terms, the gacA mutant is also impaired in growth, although the severity of this defect depends considerably on the culture medium (Whistler and Ruby 2003). In short, the gacA mutant has a pleiotropic phenotype, it is affected in a number factors known to (or likely to) affect colonization of the host, and its inability to grow to wild-type levels in the light organ is not a symbiosis-specific defect. Nonetheless, its connection to so many colonization-promoting phenotypes marks it as a global regulator of importance in the symbiosis, and the careful use of another attenuated (lysA) mutant as a control strongly suggests a critical symbiosis-specific signaling defect.
GacA is conserved in numerous and diverse γ-proteobacteria (although its orthologs have distinct names), where it controls a myriad of functions, including carbon-flow physiology and virulence (Lapouge et al. 2008). Certain themes relevant to a potential symbiotic role have emerged, including the control of factors involved in colonizing hosts and pathways for social behavior and intraspecies signaling (e.g., quorum sensing) (Lapouge et al. 2008). Also conserved is the mechanism of its associated regulatory cascade (Lapouge et al. 2008). The sensor kinase GacS is responsible for phosphorylating and thus activating the cognate response regulator GacA. Then GacA-P stimulates transcription of CsrB, regulatory RNAs that bind to CsrA, preventing it from binding target mRNAs and affecting their stability and/or translation. In V. fischeri, bioinformatic analyses revealed two such regulatory RNAs, named CsrB1 and CsrB2 (Kulkarni et al. 2006), and putative GacA-binding sites are present upstream of each of these genes. V. fischeri also possesses clear homologs of GacS and CsrA. Unpublished results from multiple laboratories suggest that, as predicted, CsrB1, CsrB2, CsrA, and GacS have regulatory functions in V. fischeri, at least some of which are interconnected. This regulatory circuit is an area of active research, particularly with respect to its effect on bioluminescence from the lux system and a global perspective of its regulon, which is likely to include genes involved in signals perceived by the host.
Important questions that remain include the signal or environmental cue perceived by GacS and whether this is present in the light-organ environment. The altered symbiotic phenotype of gacA mutants suggests the GacS/GacA system is active during colonization, although it cannot be ruled out that unphosphorylated GacA has some activity or that GacA is phosphorylated by some means other than GacS (e.g., by cross talk from another sensor kinase). The stimulus for GacS homologs in other systems has been elusive, although it may involve metabolic products of the bacteria themselves (Takeuchi et al. 2009), and could perhaps be sensed from the cytoplasmic rather than periplasmic side of GacS (Zuber et al. 2003). If so, the Gac regulon could reflect a regulatory response to symbiont physiology as it is constrained by the host environment. This possibility underscores the importance of understanding symbiont physiology and how V. fischeri metabolism is supported by the host. The V. fischeri-E. scolopes model system promises to be a rich and tractable experimental system for deciphering the widespread Gac-Csr system.
N-Acetyl d-Glucosamine Repressor NagC
The bacterial transcriptome analysis of Wier et al. (2010) described above revealed that genes involved in chitin catabolism were upregulated prior to dawn (Wier et al. 2010). This finding stimulated an interest in understanding the regulation of these genes. Specifically, Miyashiro et al. (2011) searched for regulators of the exochitinase gene, VF_1598, and found that the repressor NagC negatively controls its expression and also represses the nagA operon (which includes nagC itself, VF_0806). Loss of nagC increased expression of these and several other genes involved in the utilization of chitin or its derivatives, the monomer N-acetyl-d-glucosamine (GlcNAc) or the dimer chitobiose (GlcNAc2). Excitingly, in colonization assays, a nagC null mutant exhibited a severe defect: when the nagC mutant was used to inoculate squid, most squid remained uncolonized (6 of 15 became colonized within 48 h compared to 90% colonized with the wild-type control) (Miyashiro et al. 2011). In competition assays, the nagC mutant neither impaired wild-type colonization nor was complemented by the presence of the wild-type strain. In contrast to the nagC result, no defect was observed for a nagB mutant, which lacks a deaminase involved in releasing the amino group from GlcNAc-6P; this mutant failed to grow preferentially on GlcNAc as a carbon and nitrogen source. Thus, the symbiotic defect of the nagC mutant is unlikely to stem from altered metabolism of GlcNAc. However, the generation and evaluation of a nagC nagB double mutant would better address the symbiotic consequences of the overexpression of GlcNAc metabolism genes resulting from nagC disruption. Finally, exposure to GlcNAc during inoculation eliminated the competitive advantage of the wild type over the nagC mutant (Miyashiro et al. 2011). Together, these data indicate that NagC represses something that must be turned off for colonization to proceed normally; in the absence of NagC-mediated repression, colonization is severely impaired. It will be of great interest to determine the regulon of NagC and which regulon member(s) impair colonization when inappropriately expressed.
Possible Role for Pili in Symbiosis
Many pathogenic associations depend upon a pilus-mediated adherence of the bacteria to the host (Pizarro-Cerda and Cossart 2006). For example, type I pili play critical roles in attachment and uptake in urinary tract infections by uropathogenic E. coli (Mulvey et al. 1998; Wright et al. 2007). To date, however, a critical role for pili in the V. fischeri-E. scolopes symbiosis has yet to be determined, although several studies have investigated this possibility. Interestingly, V. fischeri contains ten loci encoding putative pili (Ruby et al. 2005). Eight loci encode putative type IV pili, the most common type of bacterial pilus (reviewed in (Burrows 2005; Pelicic 2008; Pizarro-Cerda and Cossart 2006)). Due in part to their retractability, type IV pili play a variety of important roles in other bacteria, including adherence to various surfaces and hosts, twitching motility, and secretion of a variety of factors. Intriguingly, one of the type IV pilus loci is similar to that of the V. cholerae TCP (toxin co-regulated pilus), an important virulence factor in that bacterium. Although some of the V. fischeri genes are not clustered but are spread throughout the genome, most of the TCP genes appear present (Ruby et al. 2005). One of the other pilin loci includes a gene for MshA (mannose-sensitive hemagglutin), an adhesin located at the tip of the pilus that recognizes mannose-based receptors. Intriguingly, when a mannose analog was added to seawater, it inhibited colonization by V. fischeri, suggesting that a bacteria-mannose interaction may be important during initiation (McFall-Ngai et al. 1998). Of the two non-TCP loci, one appears to encode curli, an adherence factor found in the Enterobacteriaceae (Barnhart and Chapman 2006). Interestingly, the curli locus is absent in other well-studied Vibrios, including V. cholerae, V. parahaemolyticus, and V. vulnificus (Ruby et al. 2005). Lastly, the large plasmid (pES100) of V. fischeri encodes conjugative pili (Ruby et al. 2005; Dunn et al. 2005).
Of the 10 loci, two have been characterized in some detail. One of the type IV pilus loci contains a single pilus gene (VF_A0148), encoding a PilA-like pilin protein (PilA2) predicted to be the major external subunit of the pilus (Stabb and Ruby 2003; Ruby et al. 2005). Disruption of this gene caused a small but statistically significant defect in colonization when the mutant was presented to the squid in a mixture with the wild-type strain. Potentially, the presence of one or both of the two other pilA genes compensates for the loss of VF_A0148. In support of this idea is the lack of adjacent pilus structural and assembly genes, suggesting that PilA2 gets assembled into a pilus that is associated with a distinct pilin protein (Ruby et al. 2005). An examination of the presence of pilA2 in V. fischeri isolates revealed that it is conserved in strains isolated from other light-organ symbioses, including strains from E. tasmanica (from Australia) and those from Sepiola species (from France) (Browne-Silva and Nishiguchi 2008), further suggesting that this protein may be important in bacteria-host associations. Additional genetic characterization, including the generation of mutants that lack all three pilA genes and combinations thereof, will be necessary to determine the role of pilA in attachment and colonization.
Limited characterization of the genes encoding the conjugative pilus has also been performed. These genes (VF_B38-55) are carried on the large plasmid pES100, and they are clustered with other components of a putative conjugative transfer apparatus (Dunn et al. 2005). Indeed, experiments indicated that pES100 can direct conjugative transfer of other plasmids and is almost certainly self-transmissible (Dunn et al. 2005). Efficient transfer also required the chromosomally encoded RecA protein to be present in the donor, and this requirement was not due to homologous recombination between pES100 and the plasmid it mobilized. An exciting future direction is determining whether conjugation occurs or is even induced during colonization.
Cellobiose
V. fischeri exhibits the ability to grow on cellobiose, a feature that distinguishes it from a number of other Vibrio species, including V. parahaemolyticus, V. hollisae, V. mimicus, and V. cholerae. This ability is conferred by the cel locus (VF_0603-VF_0608), consisting of three genes that comprise a phosphotransferase system (PTS) II (celABC), a putative glucokinase gene CelK, and a 6-phospho-β-glucosidase gene celG (Adin et al. 2008b). Negatively controlling this locus is a LacI-family regulator, CelI, which is encoded at the end of the operon. Loss of celI or the addition of cellobiose (but not other sugars) resulted in colonies that turned blue on the colorimetric substrate X-gal, a phenotype that depended upon an intact celG. Further investigations revealed that CelG could cleave a variety of substrates with a β-1,4 linkage and could serve both as a β-glucosidase (to cleave substrates such as cellobiose) and as a β-galactosidase (consistent with its ability to cleave X-gal); its highest activity was as a β-glucosidase, as that activity was 50-fold higher than its β-galactosidase activity. Although not a preferred substrate, the ability of CelG to cleave X-gal has implications for the use of lacZ as a reporter in V. fischeri (Adin et al. 2008b). Despite the somewhat unique ability of V. fischeri to use cellobiose, this ability was not required for colonization: competition experiments of various cel mutants with wild type revealed no competitive disadvantage of the lack of cellobiose catabolism; the exception to this was the celI mutant, which constitutive expressed the cel genes and exhibited a twofold defect in competitions with the wild-type strain for colonization. This inability of the celI mutant to compete could be due to the overexpression of the cel locus, but could not be attributed to the cellobiose catabolism, as a celI celG mutant exhibited a similar competitive defect (Adin et al. 2008b). Finally, a mutation in the unlinked gene ptsI (VF_1895/1896), which encodes the E1 component of the PTS system, exhibited a defect in cellobiose uptake as well as a severe competitive defect in colonization. However, because this strain also exhibited a growth defect, its specific role in colonization remains uncertain. These studies thus add to our knowledge and understanding of the physiology of V. fischeri important to its ability to colonize its host E. scolopes and provide a cautionary note for those who rely on lacZ as a reporter.
Other Genes
The roles in symbiosis of a number of additional genes have been investigated. Some, such as phosphoglucomutase (pgm), play significant but poorly understood roles in symbiosis, while others have been shown to make small contributions, and yet others have an impact on symbiosis that cannot be separated from their requirement for normal growth of V. fischeri. A select set of these genes is included in Table 20.1 . As the study of V. fischeri symbiosis continues and the interactions between V. fischeri and its host become better defined, the roles of these genes may become better understood.
Evolution of the V. fischeri-E. scolopes Symbiosis
Due to its experimental tractability, the V. fischeri-E. scolopes model has great potential to inform our understanding of evolutionary relationships between bacteria and their specific hosts. In the Hawaiian archipelago, at least two geographically isolated and genetically distinct populations of E. scolopes exist (Maunalua Bay and Kaneohe Bay, Oahu) (Jones et al. 2006; Kimbell et al. 2002). Furthermore, numerous related Vibrio-containing squids can be found around the world, including those of the same genus (e.g., Euprymna tasmanica from Australia and Euprymna morsei from Japan) as well as those from the distinct genus Sepiola (e.g., Sepiola affinis and Sepiola robusta from France). A wealth of questions can be addressed about symbiont specificity and evolution using phylogenetic and comparative analyses and experimental manipulation of established symbiosis models.
An early study analyzed coevolution of the bacteria and squid. Specifically, Nishiguchi et al. (1998) asked whether V. fischeri strains isolated from E. scolopes (“native” bacteria) were more successful at colonizing E. scolopes than strains isolated from other squid species (“nonnative” bacteria). In competition assays in which a mixture of strains was used to inoculate juvenile E. scolopes, the squid predominantly became colonized with native strains rather than nonnative strains (Nishiguchi et al. 1998). Furthermore, when two nonnative strains competed, the strain that was more closely related to the native symbiont was also the better colonizer of juvenile E. scolopes. Parallel results were obtained from competition of strains from E. tasmanica and E. hyllebergi (Nishiguchi 2002). These studies suggest that the bacteria have evolved to better colonize a specific host. The 1998 study also described evidence of parallel cladogenesis, as assessed using sequence divergence at two loci in related squids, the ITS (internal transcribed spacer between the 18S and 25S rRNA genes) and COI (cytochrome oxidase subunit I) and gapA (glyceraldehyde phosphate dehydrogenase) in the bacteria (Nishiguchi et al. 1998). However, a subsequent study determined that the bacterial gene sequenced was not gapA, but rather epd, and reanalysis of the data failed to support the original conclusions (Dunlap et al. 2007). Indeed, the latter study found no evidence from phylogenetic analyses for parallel evolution in either squid/bacteria or fish/bacteria symbioses. This may reflect the facultative nature of this symbiosis: V. fischeri is an extracellular symbiont acquired anew from the environment in each generation of squid.
In a more recent study, Wollenberg and Ruby investigated whether the symbionts from the two geographically separated populations of E. scolopes squid (in Maunalua Bay and Kaneohe Bay, Oahu, Hawaii) could be phenotypically distinguished (Wollenberg and Ruby 2009). Bacteria from the two populations of squid were collected over several nights and subsequently cultured to determine their phenotypes with respect to bioluminescence level, colony pigmentation, motility, growth rate, and siderophore production. Except for siderophore production, these characteristics could be used to distinguish the organisms isolated from the two squid populations in a manner that was statistically significant. These data support the idea that the squid host influences the generation of genetically distinct populations of V. fischeri (Wollenberg and Ruby 2009). A subsequent study, using four bacterial housekeeping genes (recA, mdh, katA, and pyrC), identified a monophyletic group (“group A”) of strains that could be found at a higher frequency than other strains in Maunalua squid (Wollenberg and Ruby 2012). Group A was able to outcompete non-group A strains in colonizing Maunalua squid hosts; however, this group appeared to be at a disadvantage when free-living in the Maunalua Bay environment (Wollenberg and Ruby 2012). These findings suggested a “fitness trade-off” for growth in the host versus survival in the environment.
Studies of Vibrio isolates obtained elsewhere, particularly in the Mediterranean where multiple sympatric squid species reside, suggest that they are genetically diverse (Jones et al. 2006; Nyholm and Nishiguchi 2008). Thus, factors other than host availability may influence the population structure of the bacteria (Nyholm and Nishiguchi 2008). Such factors may include water flow, which can impact the distribution of the bacteria; salinity and temperature, which may exert selective pressures on the bacteria (Jones et al. 2007; Nishiguchi 2000; Soto et al. 2009); and competition from other species (Wollenberg and Ruby 2012).
The phylogenetic tree constructed by Wollenberg and Ruby (2012) revealed that E. scolopes symbionts form a polyphyletic clade within V. fischeri (Wollenberg and Ruby 2012). These data represent an expansion of a similar data set generated by Mandel et al. (2009). The Mandel study investigated host specificity by comparing the genomes of two V. fischeri strains, one isolated from the squid (ES114) and the other isolated from the fish Monocentris japonica (MJ11); the latter strain fails to colonize squid proficiently (Mandel et al. 2009; McFall-Ngai and Ruby 1991; Schuster et al. 2010). Genomic comparisons revealed that the two strains were quite similar: 91% of the genes encoded proteins with a median amino acid identity of at least 99% (Mandel et al. 2009). Thus, there is limited diversity within this group of bacteria, unlike in other organisms like some species within the Enterobacteriaceae, which contain large, diverse genomic islands (Welch et al. 2002).
These studies also revealed the absence of the sensor kinase gene rscS from the fish symbiont. Mandel et al. (2009) proposed that the absence of rscS, required for initiation of symbiosis by ES114 (Visick and Skoufos 2001; Yip et al. 2006), could be sufficient to account for the inability of the fish symbiont to colonize E. scolopes (Mandel et al. 2009). This hypothesis was experimentally addressed by introducing rscS into the fish symbiont. This genetically modified strain became competent to colonize squid. These experiments supported the conclusion that rscS serves as a specificity factor that promotes colonization of E. scolopes by the subset of strains that contain it. Mandel et al. (2009) subsequently investigated the origin of rscS in V. fischeri strains and determined that rscS likely arose in a single horizontal acquisition event from an unknown donor prior to the introduction of V. fischeri into squid in the North Pacific Ocean. Subsequent transmission of rscS resulted in two major alleles of rscS, termed rscS A and rscS B. Squid symbionts contained rscS A, while half of the fish symbionts lacked rscS altogether. The half of the fish symbionts that contained rscS largely carried the rscS B allele; the rscS B allele was not sufficient to promote colonization of squid (Mandel et al. 2009). These data suggested that these rscS-containing fish symbionts descended from squid symbionts, and these bacteria either dispensed with or modified rscS. These experiments highlight the power of whole-genome comparisons and the utility of combining phylogenetic analyses with experimental measurements of colonization.
One major exception to the striking similarity between the ES114 and MJ11 genomes was found in the bioluminescence genes (Bose et al. 2011; Mandel et al. 2009). Although the genes and their arrangement were conserved, the proteins encoded by luxR/I and luxCDABE had only between 75% and 89.5% identity at the amino acid level, whereas most other proteins, including those whose genes flank the lux operon, were more than 95% identical. Moreover, the intergenic region between luxR and the lux operon showed a much greater divergence than intergenic regions of nearby genes (Bose et al. 2011). A comparison of the luxRI intergenic region of 18 V. fischeri strains revealed that only 104 of the 222 base-pair positions were conserved in all of the strains. When these sequences were used to generate a gene tree, two distinct clades were revealed, one of which encompassed all the strains classified as having highly visible luminescence. The second clade included all of the isolates from E. scolopes, which are non-visibly luminescent in culture, as well as isolates from E. tasmanica and E. morsei and a few planktonic isolates. To understand the significance of the diversity within this intergenic region, Bose et al. (2011) compared it to another intergenic region, that between glpA and fdhA. The latter intergenic region was conserved at 97% of the positions (289 of 298 bp). These data are consistent with the conclusion that the lux intergenic region is under different selective pressure—presumably the host environment—that has led to a relatively rapid evolution of bioluminescence genes and regulatory regions (Bose et al. 2011).
The above studies have been nicely complemented by an experimental evolution approach reported by Schuster et al. (2010). In this study, juvenile E. scolopes were exposed to two nonnative strains of V. fischeri: MJ11, the fish symbiont described above, and WH1, a free-living strain isolated from a region of the USA that does not harbor these squid. Although not native to E. scolopes, these V. fischeri isolates could colonize given a high inoculation dose and enough time. These strains then were serially passaged through 14 additional hatchling squid as follows. Every day, squid were rinsed and moved to fresh seawater. On the third day following inoculation, the seawater into which bacteria were vented by colonized animals was used as the inoculum for the next newly hatched squid. Thus, with the exception of the initial inoculation with cultured bacteria, these experiments were culture-independent and mimicked the natural cycle of the symbiosis through initiation, growth and persistence, venting, and persistence in the seawater. Importantly, this approach selected for improved traits at a number of different stages, rather than simply one, such as initiation. It was estimated that the bacteria in these experiments underwent a total of between 290 and 360 symbiotic generations.
During this limited experimental evolution, descendents of the two nonnative strains, WH1 and MJ11, exhibited clear phenotypic differences from their parents, notably with respect to luminescence output (Schuster et al. 2010). In contrast to the native symbiont ES114, which is non-visibly luminescent, WH1 and MJ11 produce high levels of visible light. Evolved derivatives of WH1 and MJ11 showed convergent evolution toward reduced light production. Specifically, in each of six independently evolved lines of MJ11, non-visibly luminescent isolates were observed; in four of these lines, all of the isolates examined were non-visibly luminescent. For WH1, four of the six lines yielded strains with decreased bioluminescence. Intriguingly, although the decrease in light production of the evolved WH1 strains was not substantial (approximately twofold) when light production was measured in vitro, the same strains exhibited about a 50-fold decrease in symbiotic (in vivo) bioluminescence per cell. These data supported the hypothesis that the decrease in light production was a meaningful change in the context of symbiosis. To test this idea further, Schuster et al. (2010) asked whether decreased luminescence was an advantage or disadvantage by exposing squid to two strains (from the same evolved population) that exhibited different luminescence levels (ancestral or decreased luminescence). They found that the strain with decreased luminescence outcompeted the one with ancestral luminescence levels, suggesting that this evolved phenotype may in fact be advantageous during the symbiotic life cycle of E. scolopes.
Together, these data indicate that decreased luminescence is one adaptation V. fischeri makes rapidly to become a proficient colonizer of the E. scolopes host. Furthermore, although it is clear that the squid select for strains that are proficient to produce light (Visick et al. 2000; Wollenberg et al. 2012), it seems likely that there is additional selection to maintain a luminescence level that is not excessive (Schuster et al. 2010). Future work that assesses the full complement of changes that occur during the experimental evolution, and a determination of which changes make important contributions to colonization proficiency, will provide great insights into the mechanisms of symbiont evolution and the selective forces that direct these processes.
Perspectives and Future of the Field
The V. fischeri-E. scolopes symbiosis is a powerful experimental model for elucidating bacteria-host interactions and the impact of bacteria on host development. One strength of the system comes from the fact that research into squid biology and how bacteria influence host development informs investigations into the roles of bacteria in symbiosis and vice versa. The result is a synergy of inquiry that has resulted in great advances in our understanding.
Another reason that work on the system has progressed so readily is the continued development of genetic tools that greatly facilitate the manipulation of the bacteria. Early work focused on the generation of V. fischeri mutants using such blunt instruments as transposon mutagenesis, gene replacement (with corresponding insertion of antibiotic resistance cassettes), and insertional mutagenesis. These tools were critical at the time and are still used to great advantage. However, it is now also possible to readily and rapidly generate in-frame deletions or incorporate targeted point mutations into the V. fischeri chromosome. Refinement of the ES114 genome database combined with whole-genome resequencing also now allows researchers to pinpoint spontaneous mutations and to track the genetic changes in evolved derivatives of the wild type. This new technology could solve old puzzles. For example, genomic sequencing of the hypermotile strains isolated by Millikan and Ruby (2002) may lead us to identify new regulators of motility and symbiosis, while sequencing the visibly bioluminescent derivative of ES114 (Dunlap et al. 1995) may uncover novel pathways of luminescence regulation. Taken together, this expanding genetic toolbox will facilitate ever more refined investigations of the roles of specific genes and their products during symbiosis. Similarly, in the future, the current construction of a comprehensive library of catalogued mutants, encompassing disruptions of nonessential genes, will provide an invaluable resource for mutant analyses.
Another critical advance has been the development of GFP both as a marker, to visualize where the bacterial cells are in the symbiosis, and as a reporter, to provide information on gene expression in symbiotic cells in real time. Our understanding of the establishment of this symbiosis stems largely from the seminal work of Nyholm et al. (2000), who first used GFP-labeled cells to visually discover aggregation and other early infection events. Subsequent work with V. fischeri expressing proteins with red fluorescence enabled the visualization of two infecting strains at once (Dunn et al. 2006) and provides a fluorescent standard against which GFP expression can be normalized (Miyashiro et al. 2010). Similarly, researchers studying the squid have pioneered a number of fluorescence-based microscopic methods to investigate processes such as apoptosis or specific proteins in the host. The transparent tissue of the light organ affords a rare opportunity to use fluorescence-based methods to visualize symbiotic processes in intact tissues, and the development of other fluorophores, destabilized GFP derivatives, and various animal-tissue stains should continue to be exploited to great effect.
An increased basic understanding of V. fischeri genetic processes has both provided tools and led to new intriguing questions. For example, the identification of a replication origin from a plasmid endogenous to V. fischeri permitted the construction of stable shuttle plasmids that could be readily used for a variety of experiments such as complementation or the strain-tagging with GFP, described above. The description of native V. fischeri plasmids has also raised the possibility that genetic information is being exchanged through conjugation between different strains during symbiosis. Similarly, the development of natural transformation as a tool not only provided another method for genetic manipulation of V. fischeri but also suggested the possibility of DNA uptake and recombination during symbiosis (Pollack-Berti et al. 2010). Taken together, one wonders whether future investigations will find that the packed confines of the light organ could be a hot spot for horizontal gene transfer.
As in many bacterial systems over the last decades, genomic approaches have transformed our lens into this symbiosis. The availability of the genome sequence of V. fischeri strain ES114 (Fig. 20.10 ), and subsequently that of the fish symbiont strain MJ11, made it possible to use bioinformatics to make predictions about the importance of genes in the symbiosis and to readily clone wild-type and mutant alleles of specific genes. Comparative genomic studies will become even more important as the sequences of additional genomes become available, and as the studies of symbiotic associations of V. fischeri with other squid species (as well as studies of other bacteria-light-organ symbioses) become more developed (Guerrero-Ferreira and Nishiguchi 2009; Nyholm and Nishiguchi 2008). Likewise, the squid EST and proteome databases have already provided insight into how the host detects and responds to V. fischeri, and future work is likely to expand these resources. Additional molecular technology grounded in genomics, bioinformatics, and high-throughput methods—from arrays to RNAseq to single-cell analyses—holds great promise for elucidating bacterial responses to the colonization process (and the corresponding responses of the squid). We anticipate that the next decade will see a great expansion in the availability and productive use of these approaches.
In some cases, knowledge from the system has been slow to impact the conduct of current research. For example, it has been known for a long time that juvenile squid have six pores and six distinct crypts, and we now know that it is rare for those crypts to be highly colonized by multiple strains: in competition experiments using two differently labeled strains, most crypts contain one or the other strain but usually not both (Fig. 20.4 ) (Dunn et al. 2006). Furthermore, the extent to which the different crypts expel their symbionts in the morning varies. For the most part, this information has not yet impacted our straightforward interpretation of competition experiments: if more cells of one strain are obtained following a competitive colonization experiment, then we conclude that that strain had a colonization advantage. But, did this advantage flow from superior entry into multiple crypts, superior binding within a single crypt relative to another strain, being lucky enough to colonize the right crypt, or some other reason? Follow-up experiments that visualize the colonized animals using fluorescently labeled strains will provide better insights into the requirements for a particular gene/trait.
In the early days of investigating this symbiosis, insights from other systems—and especially pathogenic systems—were used to develop models for how V. fischeri and E. scolopes interact. For example, it made sense to ask whether flagella were important for colonization, as this was the case for many pathogenic associations. As the field has developed, however, our studies have begun to provide new insights that can inform the work of others. Notably, the identification of H-NOX as a NO sensor and NO as a potential signal has clear implications for pathogenic associations (Wang et al. 2010a). Similarly, the novel nuclear localization of EsPGRP1 and the role of the symbionts in changing the localization of receptors for MAMPS have potential to impact studies of the immune response (Troll et al. 2009a, b). A third example lies in the paucity of good animal models for studying bacterial biofilms; the squid symbiosis makes a robust model, as the parallels between phenotypes observed in culture and during symbiosis are very strong, and thus the potential for informing the biofilm field is high (Morris et al. 2011; Yip et al. 2006).
Many questions remain. Although much is known about conditions and molecules that influence the activities of these organisms and their interactions, including TCT, PG, pheromones, and light itself, numerous bacterial regulators have been identified for which the signal transduction cascade remains to be elucidated. For example, the sensor kinase RscS was identified over 10 years ago, but the signal to which it responds during symbiosis remains elusive. Whether V. fischeri recognizes a chemotactic signal to guide it into the light organ and what drives the loss of flagella in colonizing bacteria are among the longstanding intriguing questions about colonization. Indeed, we have yet to truly understand how specificity in this symbiosis is achieved; perhaps there is no one single factor but a multitude of them, such as the ability to form a biofilm, sense a chemotactic signal, resist host defenses, and induce the right set of responses, that together result in the successful symbiont. The opportunity to be the sole occupant of a “privileged” niche is a powerful incentive, and V. fischeri has clearly evolved to make use of this prime opportunity for self-advancement. With the increasing sophistication of the tools, approaches, and knowledge of the system, it seems likely that many of the current secrets shortly will be revealed, but also that many more will present themselves.
References
Adin DM, Engle JT, Goldman WE, McFall-Ngai MJ, Stabb EV (2009) Mutations in ampG and lytic transglycosylase genes affect the net release of peptidoglycan monomers from Vibrio fischeri. J Bacteriol 191:2012–2022
Adin DM, Phillips NJ, Gibson BW, Apicella MA, Ruby EG, McFall-Ngai MJ, Hall DB, Stabb EV (2008a) Characterization of htrB and msbB mutants of the light organ symbiont Vibrio fischeri. Appl Environ Microbiol 74:633–644
Adin DM, Visick KL, Stabb EV (2008b) Identification of a cellobiose utilization gene cluster with cryptic β-galactosidase activity in Vibrio fischeri. Appl Environ Microbiol 74:4059–4069
Aeckersberg F, Lupp C, Feliciano B, Ruby EG (2001) Vibrio fischeri outer membrane protein OmpU plays a role in normal symbiotic colonization. J Bacteriol 183:6590–6597
Alcaide E (2003) Numerical taxonomy of Vibrionaceae isolated from cultured amberjack (Seriola dumerili) and surrounding water. Curr Microbiol 46:184–189
Altura MA, Stabb E, Goldman W, Apicella M, McFall-Ngai MJ (2011) Attenuation of host NO production by MAMPs potentiates development of the host in the squid-vibrio symbiosis. Cell Microbiol 13:527–537
Anderson RC, Mather JA (1996) Escape responses of Euprymna scolopes Berry, 1911 (Cephalopoda: Sepiolidae). J Moll Stud 62:543–545
Antunes LC, Ferreira RB, Lostroh CP, Greenberg EP (2008) A mutational analysis defines Vibrio fischeri LuxR binding sites. J Bacteriol 190:4392–4397
Antunes LC, Schaefer AL, Ferreira RB, Qin N, Stevens AM, Ruby EG, Greenberg EP (2007) Transcriptome analysis of the Vibrio fischeri LuxR-LuxI regulon. J Bacteriol 189:8387–8391
Arcondeguy T, Jack R, Merrick M (2001) P(II) signal transduction proteins, pivotal players in microbial nitrogen control. Microbiol Mol Biol Rev: MMBR 65:80–105
Baker MD, Wolanin PM, Stock JB (2006) Signal transduction in bacterial chemotaxis. BioEssays 28:9–22
Barnhart MM, Chapman MR (2006) Curli biogenesis and function. Annu Rev Microbiol 60:131–147
Berry SS (1912) The Cephalopoda of the Hawaiian islands. In: Bulletin of the United States Bureau of Fisheries. Government Printing Office, Washington, pp 255–362
Boettcher KJ, Ruby EG (1990) Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J Bacteriol 172:3701–3706
Boettcher KJ, Ruby EG (1994) Occurrence of plasmid DNA in the sepiolid squid symbiont Vibrio fischeri. Curr Microbiol 29:279–286
Boettcher KJ, Ruby EG, McFall-Ngai MJ (1996) Bioluminescence in the symbiotic squid Euprymna scolopes is controlled by a daily biological rhythm. J Comp Physiol 179:65–73
Bose JL, Kim U, Bartkowski W, Gunsalus RP, Overley AM, Lyell NL, Visick KL, Stabb EV (2007) Bioluminescence in Vibrio fischeri is controlled by the redox-responsive regulator ArcA. Mol Microbiol 65:538–553
Bose JL, Rosenberg CS, Stabb EV (2008) Effects of luxCDABEG induction in Vibrio fischeri: enhancement of symbiotic colonization and conditional attenuation of growth in culture. Arch Microbiol 190(2):169–183
Bose JL, Wollenberg MS, Colton DM, Mandel MJ, Septer AN, Dunn AK, Stabb EV (2011) Contribution of rapid evolution of the luxR-luxI intergenic region to the diverse bioluminescence outputs of Vibrio fischeri strains isolated from different environments. Appl Environ Microbiol 77:2445–2457
Bourgois JJ, Sluse FE, Baguet F, Mallefet J (2001) Kinetics of light emission and oxygen consumption by bioluminescent bacteria. J Bioenerg Biomembr 33:353–363
Boylan M, Miyamoto C, Wall L, Grahm A, Meighen E (1989) Lux C, D, and E genes of the Vibrio fischeri luminescence operon code for the reductase, transferase, and synthetase enzymes involved in aldehyde biosyntheses. Photochem Photobiol 49:681–688
Browne-Silva J, Nishiguchi MK (2008) Gene sequences of the pil operon reveal relationships between symbiotic strains of Vibrio fischeri. Int J Syst Evol Microbiol 58:1292–1299
Buck M, Gallegos M-T, Studholme DJ, Guo Y, Gralla JD (2000) The bacterial enhancer-dependent σ54 (σN) transcription factor. J Bacteriol 182:4129–4136
Burrows LL (2005) Weapons of mass retraction. Mol Microbiol 57:878–888
Callahan SM, Dunlap PV (2000) LuxR- and acyl-homoserine-lactone-controlled non-lux genes define a quorum sensing regulon in Vibrio fischeri. J Bacteriol 182:2811–2822
Castillo MG, Goodson MS, McFall-Ngai M (2009) Identification and molecular characterization of a complement C3 molecule in a lophotrochozoan, the Hawaiian bobtail squid Euprymna scolopes. Dev Comp Immunol 33:69–76
Chen X, Schauder S, Potier N, Van Dorsselaer A, Pelczer I, Bassler BL, Hughson FM (2002) Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415:545–549
Chevance FF, Hughes KT (2008) Coordinating assembly of a bacterial macromolecular machine. Nat Rev Microbiol 6:455–465
Chun CK, Scheetz TE, Bonaldo Mde F, Brown B, Clemens A, Crookes-Goodson WJ, Crouch K, DeMartini T, Eyestone M, Goodson MS, Janssens B, Kimbell JL, Koropatnick TA, Kucaba T, Smith C, Stewart JJ, Tong D, Troll JV, Webster S, Winhall-Rice J, Yap C, Casavant TL, McFall-Ngai MJ, Soares MB (2006) An annotated cDNA library of juvenile Euprymna scolopes with and without colonization by the symbiont Vibrio fischeri. BMC Genomics 7:154
Chun CK, Troll JV, Koroleva I, Brown B, Manzella L, Snir E, Almabrazi H, Scheetz TE, Bonaldo Mde F, Casavant TL, Soares MB, Ruby EG, McFall-Ngai MJ (2008) Effects of colonization, luminescence, and autoinducer on host transcription during development of the squid-vibrio association. Proc Natl Acad Sci USA 105:11323–11328
Claes MF, Dunlap PV (2000) Aposymbiotic culture of the sepiolid squid Euprymna scolopes: role of the symbiotic bacterium Vibrio fischeri in host animal growth, development, and light organ morphogenesis. J Exp Zool 286:280–296
Correa NE, Lauriano CM, McGee R, Klose KE (2000) Phosphorylation of the flagellar regulatory protein FlrC is necessary for Vibrio cholerae motility and enhanced colonization. Mol Microbiol 35:743–755
Correa NE, Peng F, Klose KE (2005) Roles of the regulatory proteins FlhF and FlhG in the Vibrio cholerae flagellar transcription hierarchy. J Bacteriol 187:6324–6332
Czyz A, Plata K, Wegrzyn G (2003) Stimulation of DNA repair as an evolutionary drive for bacterial luminescence. Lumin: J Biol Chem Lumin 18:140–144
Darnell CL, Hussa EA, Visick KL (2008) The putative hybrid sensor kinase SypF coordinates biofilm formation in Vibrio fischeri by acting upstream of two response regulators, SypG and VpsR. J Bacteriol 190:4941–4950
Davidson SK, Koropatnick TA, Kossmehl R, Sycuro L, McFall-Ngai MJ (2004) NO means ‘yes’ in the squid-vibrio symbiosis: nitric oxide (NO) during the initial stages of a beneficial association. Cell Microbiol 6:1139–1151
Deloney-Marino CR, Visick KL (2012) Role for cheR of Vibrio fischeri in the Vibrio-squid symbiosis. Can J Microbiol 58:29–38
DeLoney-Marino CR, Wolfe AJ, Visick KL (2003) Chemoattraction of Vibrio fischeri to serine, nucleosides and N-acetylneuraminic acid, a component of squid light organ mucus. Appl Environ Microbiol 69:7527–7530
DeLoney CR, Bartley TM, Visick KL (2002) Role for phosphoglucomutase in Vibrio fischeri-Euprymna scolopes symbiosis. J Bacteriol 184:5121–5129
Doino JA, McFall-Ngai MJ (1995) A transient exposure to symbiosis-competent bacteria induces light organ morphogenesis in the host squid. Biol Bull 189:347–355
Doino Lemus J, McFall-Ngai MJ (2000) Alterations in the proteome of the Euprymna scolopes light organ in response to symbiotic Vibrio fischeri. Appl Environ Microbiol 66:4091–4097
Dunlap PV, Ast JC, Kimura S, Fukui A, Yoshino T, Endo H (2007) Phylogenetic analysis of host-symbiont specificity and codivergence in bioluminescent symbioses. Cladistics 23:507–532
Dunlap PV, Greenberg EP (1985) Control of Vibrio fischeri luminescence gene expression in Escherichia coli by cyclic AMP and cyclic AMP receptor protein. J Bacteriol 164:45–50
Dunlap PV, Greenberg EP (1988) Control of Vibrio fischeri lux gene transcription by a cyclic AMP receptor protein-LuxR protein regulatory circuit. J Bacteriol 170:4040–4046
Dunlap PV, Kita-Tsukamoto K, Waterbury JB, Callahan SM (1995) Isolation and characterization of a visibly luminous variant of Vibrio fischeri strain ES114 from the sepiolid squid Euprymna scolopes. Arch Microbiol 164:194–202
Dunlap PV, Ray JM (1989) Requirement for autoinducer in transcriptional negative autoregulation of the Vibrio fischeri luxR gene in Escherichia coli. J Bacteriol 171:3549–3552
Dunn AK, Karr EA, Wang Y, Batton AR, Ruby EG, Stabb EV (2010) The alternative oxidase (AOX) gene in Vibrio fischeri is controlled by NsrR and upregulated in response to nitric oxide. Mol Microbiol 77:44–55
Dunn AK, Martin MO, Stabb E (2005) Characterization of pES213, a small mobilizable plasmid from Vibrio fischeri. Plasmid 54:114–134
Dunn AK, Millikan DS, Adin DM, Bose JL, Stabb EV (2006) New rfp- and pES213-derived tools for analyzing symbiotic Vibrio fischeri reveal patterns of infection and lux expression in situ. Appl Environ Microbiol 72:802–810
Dunn AK, Stabb EV (2007) Beyond quorum sensing: the complexities of prokaryotic parliamentary procedures. Anal Bioanal Chem 387:391–398
Dunn AK, Stabb EV (2008a) Genetic analysis of trimethylamine N-oxide reductases in the light organ symbiont Vibrio fischeri ES114. J Bacteriol 190:5814–5823
Dunn AK, Stabb EV (2008b) The twin arginine translocation system contributes to symbiotic colonization of Euprymna scolopes by Vibrio fischeri. FEMS Microbiol Lett 279:251–258
Duperthuy M, Schmitt P, Garzon E, Caro A, Rosa RD, Le Roux F, Lautredou-Audouy N, Got P, Romestand B, de Lorgeril J, Kieffer-Jaquinod S, Bachere E, Destoumieux-Garzon D (2011) Use of OmpU porins for attachment and invasion of Crassostrea gigas immune cells by the oyster pathogen Vibrio splendidus. Proc Natl Acad Sci USA 108:2993–2998
Egland KA, Greenberg EP (1999) Quorum sensing in Vibrio fischeri: elements of the luxl promoter. Mol Microbiol 31:1197–1204
Egland KA, Greenberg EP (2000) Conversion of the Vibrio fischeri transcriptional activator, LuxR, to a repressor. J Bacteriol 182:805–811
Engebrecht J, Nealson K, Silverman M (1983) Bacterial bioluminescence: isolation and genetic analysis of functions from Vibrio fischeri. Cell 32:773–781
Engebrecht J, Silverman M (1984) Identification of genes and gene products necessary for bacterial bioluminescence. Proc Natl Acad Sci USA 81:4154–4158
Fidopiastis PM, Miyamoto CM, Jobling MG, Meighen EA, Ruby EG (2002) LitR, a new transcriptional activator in Vibrio fischeri, regulates luminescence and symbiotic light organ colonization. Mol Microbiol 45:131–143
Fidopiastis PM, von Boletzky S, Ruby EG (1998) A new niche for Vibrio logei, the predominant light organ symbiont of squids in the genus Sepiola. J Bacteriol 180:59–64
Fidopiastis PM, Rader BA, Gerling DG, Gutierrez NA, Watkins KH, Frey MW, Nyholm SV, Whistler CA (2012) Characterization of a Vibrio fischeri aminopeptidase and evidence for its influence on an early stage of squid colonization. J Bacteriol 194:3995–4002
Fitzgerald JM (1977) Classification of luminous bacteria from the light organ of the Australian pinecone fish, Cleidopus gloriamaris. Arch Microbiol 112:153–156
Foster JS, Apicella MA, McFall-Ngai MJ (2000) Vibrio fischeri lipopolysaccharide induces developmental apoptosis, but not complete morphogenesis, of the Euprymna scolopes symbiotic light organ. Dev Biol 226:242–254
Foster JS, McFall-Ngai MJ (1998) Induction of apoptosis by cooperative bacteria in the morphogenesis of host epithelial tissues. Dev Genes Evol 208:295–303
Friedrich WF, Greenberg EP (1983) Glucose repression of luminescence and luciferase in Vibrio fischeri. Arch Microbiol 134:87–91
Fuqua C, Winans SC, Greenberg EP (1996) Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu Rev Microbiol 50:727–751
Fuqua WC, Winans SC, Greenberg EP (1994) Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol 176:269–275
Garcia-Amado MA, Bozo-Hurtado L, Astor Y, Suarez P, Chistoserdov A (2011) Denaturing gradient gel electrophoresis analyses of the vertical distribution and diversity of Vibrio spp. populations in the Cariaco Basin. FEMS Microbiol Ecol 77:347–356
Goodson MS, Kojadinovic M, Troll JV, Scheetz TE, Casavant TL, Soares MB, McFall-Ngai MJ (2005) Identifying components of the NF-κB pathway in the beneficial Euprymna scolopes-Vibrio fischeri light organ symbiosis. Appl Environ Microbiol 71:6934–6946
Graf J, Dunlap PV, Ruby EG (1994) Effect of transposon-induced motility mutations on colonization of the host light organ by Vibrio fischeri. J Bacteriol 176:6986–6991
Graf J, Ruby EG (1998) Host-derived amino acids support the proliferation of symbiotic bacteria. Proc Natl Acad Sci USA 95:1818–1822
Graf J, Ruby EG (2000) Novel effects of a transposon insertion in the Vibrio fischeri glnD gene: defects in iron uptake and symbiotic persistence in addition to nitrogen utilization. Mol Microbiol 37:168–179
Gray KM, Greenberg EP (1992a) Physical and functional maps of the luminescence gene cluster in an autoinducer-deficient Vibrio fischeri strain isolated from a squid light organ. J Bacteriol 174:4384–4390
Gray KM, Greenberg EP (1992b) Sequencing and analysis of luxR and luxI, the luminescence regulatory genes from the squid light organ symbiont Vibrio fischeri ES114. Mol Mar Biol Biotechnol 1:414–419
Guerrero-Ferreira RC, Nishiguchi MK (2009) Ultrastructure of light organs of loliginid squids and their bacterial symbionts: a novel model system for the study of marine symbioses. Vie Milieu Paris 59:307–313
Hanlon RT, Claes MF, Ashcraft SE, Dunlap PV (1997) Laboratory culture of the sepiolid squid Euprymna scolopes: a model system for bacteria-animal symbiosis. Biol Bull 192:364–374
Hanzelka BL, Parsek MR, Val DL, Dunlap PV, Cronan JE Jr, Greenberg EP (1999) Acylhomoserine lactone synthase activity of the Vibrio fischeri AinS protein. J Bacteriol 181:5766–5770
Hastings JW, Nealson KH (1977) Bacterial bioluminescence. Annu Rev Microbiol 31:549–595
Hastings JW, Riley WH, Massa J (1965) The purification, properties, and chemiluminescent quantum yield of bacterial luciferase. J Biol Chem 240:1473–1481
Haygood MG, Nealson KH (1985) Mechanisms of iron regulation of luminescence in Vibrio fischeri. J Bacteriol 162:209–216
Heath-Heckman EA, McFall-Ngai MJ (2011) The occurrence of chitin in the hemocytes of invertebrates. Zoology (Jena) 114:191–198
Hussa EA, Darnell CL, Visick KL (2008) RscS functions upstream of SypG to control the syp locus and biofilm formation in Vibrio fischeri. J Bacteriol 190:4576–4583
Hussa EA, O’Shea TM, Darnell CL, Ruby EG, Visick KL (2007) Two-component response regulators of Vibrio fischeri: identification, mutagenesis and characterization. J Bacteriol 189:5825–5838
Jones BW, Lopez JE, Huttenburg J, Nishiguchi MK (2006) Population structure between environmentally transmitted vibrios and bobtail squids using nested clade analysis. Mol Ecol 15:4317–4329
Jones BW, Maruyama A, Ouverney CC, Nishiguchi MK (2007) Spatial and temporal distribution of the vibrionaceae in coastal waters of Hawaii, Australia, and France. Microb Ecol 54:314–323
Jones BW, Nishiguchi MK (2004) Counterillumination in the Hawaiian bobtail squid, Euprymna scolopes Berry (Mollusca: Cephalopoda). Mar Biol 144:1151–1155
Kaplan HB, Greenberg EP (1985) Diffusion of autoinducer is involved in regulation of the Vibrio fischeri luminescence system. J Bacteriol 163:1210–1214
Keynan A, Hastings JW (1961) The isolation and characterization of dark mutants of luminous bacteria. Biol Bull 121:375
Kimbell JR, McFall-Ngai MJ (2003) The squid-Vibrio symbiosis: From demes to genes. Integr Comp Biol 43:254–260
Kimbell JR, McFall-Ngai MJ (2004) Symbiont-induced changes in host actin during the onset of a beneficial animal-bacterial association. Appl Environ Microbiol 70:1434–1441
Kimbell JR, McFall-Ngai MJ, Roderick GK (2002) Two genetically distinct populations of bobtail squid, Euprymna scolopes, exist on the island of Oahu. Pacific Sci 56:347–355
Koropatnick TA, Engle JT, Apicella MA, Stabb EV, Goldman WE, McFall-Ngai MJ (2004) Microbial factor-mediated development in a host-bacterial mutualism. Science 306:1186–1187
Koropatnick TA, Kimbell JR, McFall-Ngai MJ (2007) Responses of host hemocytes during the initiation of the squid-Vibrio symbiosis. Biol Bull 212:29–39
Krasity BC, Troll JV, Weiss JP, McFall-Ngai MJ (2011) LBP/BPI proteins and their relatives: conservation over evolution and roles in mutualism. Biochem Soc Trans 39:1039–1044
Kulkarni PR, Cui X, Williams JW, Stevens AM, Kulkarni RV (2006) Prediction of CsrA-regulating small RNAs in bacteria and their experimental verification in Vibrio fischeri. Nucleic Acids Res 34:3361–3369
Kuo A, Blough NV, Dunlap PV (1994) Multiple N-acyl-L-homoserine lactone autoinducers of luminescence in the marine symbiotic bacterium Vibrio fischeri. J Bacteriol 176:7558–7565
Kuo A, Callahan SM, Dunlap PV (1996) Modulation of luminescence operon expression by N-octanoyl-L-homoserine lactone in ainS mutants of Vibrio fischeri. J Bacteriol 178:971–976
Lamarcq LH, McFall-Ngai MJ (1998) Induction of a gradual, reversible morphogenesis of its host’s epithelial brush border by Vibrio fischeri. Infect Immun 66:777–785
Lamas J, Anadon R, Devesa S, Toranzo AE (1990) Visceral neoplasia and epidermal papillomas in cultured turbot Scophthalmus maximus. Dis Aquat Organ 8:179–187
Lapouge K, Schubert M, Allain FH, Haas D (2008) Gac/Rsm signal transduction pathway of gamma-proteobacteria: from RNA recognition to regulation of social behaviour. Mol Microbiol 67:241–253
Lee K-H, Ruby EG (1992) Detection of the light organ symbiont, Vibrio fischeri, in Hawaiian seawater by using lux gene probes. Appl Environ Microbiol 58:942–947
Lee K-H, Ruby EG (1994a) Competition between Vibrio fischeri strains during initiation and maintenance of a light organ symbiosis. J Bacteriol 176:1985–1991
Lee K-H, Ruby EG (1994b) Effect of the squid host on the abundance and distribution of symbiotic Vibrio fischeri in nature. Appl Environ Microbiol 60:1565–1571
Liu X, Parales RE (2008) Chemotaxis of Escherichia coli to pyrimidines: a new role for the signal transducer Tap. J Bacteriol 190:972–979
Liu X, Wood PL, Parales JV, Parales RE (2009) Chemotaxis to pyrimidines and identification of a cytosine chemoreceptor in Pseudomonas putida. J Bacteriol 191:2909–2916
Lupp C, Hancock RE, Ruby EG (2002) The Vibrio fischeri sapABCDF locus is required for normal growth, both in culture and in symbiosis. Arch Microbiol 179:57–65
Lupp C, Ruby EG (2004) Vibrio fischeri LuxS and AinS: Comparative study of two signal synthases. J Bacteriol 186:3873–3881
Lupp C, Ruby EG (2005) Vibrio fischeri utilizes two quorum-sensing systems for the regulation of early and late colonization factors. J Bacteriol 187:3620–3629
Lupp C, Urbanowski M, Greenberg EP, Ruby EG (2003) The Vibrio fischeri quorum-sensing systems ain and lux sequentially induce luminescence gene expression and are important for persistence in the squid host. Mol Microbiol 50:319–331
Lyell NL, Dunn AK, Bose JL, Stabb EV (2010) Bright mutants of Vibrio fischeri ES114 reveal conditions and regulators that control bioluminescence and expression of the lux operon. J Bacteriol 192:5103–5114
Mandel MJ, Schaefer AL, Brennan CA, Heath-Heckman EA, Deloney-Marino CR, McFall-Ngai MJ, Ruby EG (2012) Squid-derived chitin oligosaccharides are a chemotactic signal during colonization by Vibrio fischeri. Appl Environ Microbiol 78:4620–4626
Mandel MJ, Wollenberg MS, Stabb EV, Visick KL, Ruby EG (2009) A single regulatory gene is sufficient to alter bacterial host range. Nature 458:215–218
McCann J, Stabb EV, Millikan DS, Ruby EG (2003) Population dynamics of Vibrio fischeri during infection of Euprymna scolopes. Appl Environ Microbiol 69:5928–5934
McFall-Ngai M, Brennan C, Weis V, Lamarcq L (1998) Mannose adhesin-glycan interactions in the Euprymna scolopes-Vibrio fischeri symbiosis. In: Le Gal Y, Halvorson HO (eds) New developments in marine biotechnology. Plenum Press, New York, pp 273–276
McFall-Ngai M, Heath-Heckman EA, Gillette AA, Peyer SM, Harvie EA (2011) The secret languages of coevolved symbioses: Insights from the Euprymna scolopes-Vibrio fischeri symbiosis. Semin Immunol 24:3–8
McFall-Ngai M, Montgomery MK (1990) The anatomy and morphology of the adult bacterial light organ of Euprymna scolopes Berry (Cephalopoda:Sepiolidae). Biol Bull 179:332–339
McFall-Ngai M, Nyholm SV, Castillo MG (2010) The role of the immune system in the initiation and persistence of the Euprymna scolopes–Vibrio fischeri symbiosis. Semin Immunol 22:48–53
McFall-Ngai MJ (1999) Consequences of evolving with bacterial symbionts: insights from the squid-vibrio associations. Annu Rev Ecol Syst 30:235–256
McFall-Ngai MJ, Ruby EG (1991) Symbiont recognition and subsequent morphogenesis as early events in an animal-bacterial mutualism. Science 254:1491–1494
McFall-Ngai MJ, Ruby EG (1998) Sepiolids and vibrios: when first they meet. Reciprocal interactions between host and symbiont lead to the creation of a complex light-emitting organ. BioScience 48:257–265
Millikan DS, Ruby EG (2002) Alterations in Vibrio fischeri motility correlate with a delay in symbiosis initiation and are associated with additional symbiotic colonization defects. Appl Environ Microbiol 68:2519–2528
Millikan DS, Ruby EG (2003) FlrA, a σ54-dependent transcriptional activator in Vibrio fischeri, is required for motility and symbiotic light-organ colonization. J Bacteriol 185:3547–3557
Millikan DS, Ruby EG (2004) Vibrio fischeri flagellin A is essential for normal motility and for symbiotic competence during initial squid light organ colonization. J Bacteriol 186:4315–4325
Miyamoto CM, Lin YH, Meighen EA (2000) Control of bioluminescence in Vibrio fischeri by the LuxO signal response regulator. Mol Microbiol 36:594–607
Miyashiro T, Klein W, Oehlert D, Cao X, Schwartzman J, Ruby EG (2011) The N-acetyl-d-glucosamine repressor NagC of Vibrio fischeri facilitates colonization of Euprymna scolopes. Mol Microbiol 82:894–903
Miyashiro T, Wollenberg MS, Cao X, Oehlert D, Ruby EG (2010) A single qrr gene is necessary and sufficient for LuxO-mediated regulation in Vibrio fischeri. Mol Microbiol 77:1556–1567
Montes M, Farto R, Perez MJ, Armada SP, Nieto TP (2006) Genotypic diversity of Vibrio isolates associated with turbot (Scophthalmus maximus) culture. Res Microbiol 157:487–495
Montgomery MK, McFall-Ngai M (1993) Embryonic development of the light organ of the sepiolid squid Euprymna scolopes Berry. Biol Bull 184:296–308
Montgomery MK, McFall-Ngai M (1994) Bacterial symbionts induce host organ morphogenesis during early postembryonic development of the squid Euprymna scolopes. Development 120:1719–1729
Morris AR, Darnell CL, Visick KL (2011) Inactivation of a novel response regulator is necessary for biofilm formation and host colonization by Vibrio fischeri. Mol Microbiol 82:114–130
Morris AR, Visick KL (2010) Control of biofilm formation and colonization in Vibrio fischeri: a role for partner switching? Environ Microbiol 12:2051–2059
Moynihan M (1983) Notes on the behavior of Euprymna scolopes (Cephalopoda: Sepiolidae). Behaviour 85:25–41
Mulvey MA, Lopez-Boado YS, Wilson CL, Roth R, Parks WC, Heuser J, Hultgren SJ (1998) Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science 282:1494–1497
Nairz M, Schroll A, Sonnweber T, Weiss G (2010) The struggle for iron - a metal at the host-pathogen interface. Cell Microbiol 12:1691–1702
Nijvipakul S, Wongratana J, Suadee C, Entsch B, Ballou DP, Chaiyen P (2008) LuxG is a functioning flavin reductase for bacterial luminescence. J Bacteriol 190:1531–1538
Nishiguchi MK (2000) Temperature affects species distribution in symbiotic populations of Vibrio spp. Appl Environ Microbiol 66:3550–3555
Nishiguchi MK (2002) Host-symbiont recognition in the environmentally transmitted sepiolid squid-Vibrio mutualism. Microb Ecol 44:10–18
Nishiguchi MK, Ruby EG, McFall-Ngai MJ (1998) Competitive dominance among strains of luminous bacteria provides an unusual form of evidence for parallel evolution in sepiolid squid-vibrio symbioses. Appl Environ Microbiol 64:3209–3213
Nyholm SV, Deplancke B, Gaskins HR, Apicella MA, McFall-Ngai MJ (2002) Roles of Vibrio fischeri and nonsymbiotic bacteria in the dynamics of mucus secretion during symbiont colonization of the Euprymna scolopes light organ. Appl Environ Microbiol 68:5113–5122
Nyholm SV, McFall-Ngai MJ (1998) Sampling the light-organ microenvironment of Euprymna scolopes: description of a population of host cells in association with the bacterial symbiont Vibrio fischeri. Biol Bull 195:89–97
Nyholm SV, McFall-Ngai MJ (2003) Dominance of Vibrio fischeri in secreted mucus outside the light organ of Euprymna scolopes: the first site of symbiont specificity. Appl Environ Microbiol 69:3932–3937
Nyholm SV, McFall-Ngai MJ (2004) The winnowing: establishing the squid-Vibrio symbiosis. Nat Rev Microbiol 2:632–642
Nyholm SV, Nishiguchi MK (2008) The evolutionary ecology of a sepiolid squid-Vibrio association: from cell to environment. Vie Milieu Paris 58:175–184
Nyholm SV, Stabb EV, Ruby EG, McFall-Ngai MJ (2000) Establishment of an animal-bacterial association: recruiting symbiotic vibrios from the environment. Proc Natl Acad Sci USA 97:10231–10235
Nyholm SV, Stewart JJ, Ruby EG, McFall-Ngai MJ (2009) Recognition between symbiotic Vibrio fischeri and the haemocytes of Euprymna scolopes. Environ Microbiol 11:483–493
O’Shea TM, DeLoney-Marino CR, Shibata S, Aizawa S-I, Wolfe AJ, Visick KL (2005) Magnesium promotes flagellation of Vibrio fischeri. J Bacteriol 187:2058–2065
O’Shea TM, Klein AH, Geszvain K, Wolfe AJ, Visick KL (2006) Diguanylate cyclases control magnesium-dependent motility of Vibrio fischeri. J Bacteriol 188:8196–8205
Ochman H, Moran NA (2001) Genes lost and genes found: evolution of bacterial pathogenesis and symbiosis. Science 292:1096–1099
Orndorff SA, Colwell RR (1980) Distribution and identification of luminous bacteria from the Sargasso Sea. Appl Environ Microbiol 39:983–987
Pelicic V (2008) Type IV pili: e pluribus unum? Mol Microbiol 68:827–837
Phillips NJ, Adin DM, Stabb EV, McFall-Ngai MJ, Apicella MA, Gibson BW (2011) The lipid A from Vibrio fischeri lipopolysaccharide: a unique structure bearing a phosphoglycerol moiety. J Biol Chem 286:21203–21219
Pizarro-Cerda J, Cossart P (2006) Bacterial adhesion and entry into host cells. Cell 124:715–727
Pollack-Berti A, Wollenberg MS, Ruby EG (2010) Natural transformation of Vibrio fischeri requires tfoX and tfoY. Environ Microbiol 12:2302–2311
Pooley DT, Larsson J, Jones G, Rayner-Brandes MH, Lloyd D, Gibson C, Stewart WR (2004) Continuous culture of photobacterium. Biosens Bioelectron 19:1457–1463
Prouty MG, Correa NE, Klose KE (2001) The novel σ54- and σ28-dependent flagellar gene transcription hierarchy of Vibrio cholerae. Mol Microbiol 39:1595–1609
Ramesh A, Venugopalan VK (1989) Response of enteric luminous bacteria to environmental conditions in the gut of the fish. J Appl Bacteriol 66:529–533
Ruby EG, Asato LM (1993) Growth and flagellation of Vibrio fischeri during initiation of the sepiolid squid light organ symbiosis. Arch Microbiol 159:160–167
Ruby EG, Greenberg EP, Hastings JW (1980) Planktonic marine luminous bacteria: species distribution in the water column. Appl Environ Microbiol 39:302–306
Ruby EG, Morin JG (1979) Luminous enteric bacteria of marine fishes: a study of their distribution, densities, and dispersion. Appl Environ Microbiol 38:406–411
Ruby EG, Nealson KH (1976) Symbiotic association of Photobacterium fischeri with the marine luminous fish Monocentris japonica: a model of symbiosis based on bacterial studies. Biol Bull 151:574–586
Ruby EG, Urbanowski M, Campbell J, Dunn A, Faini M, Gunsalus R, Lostroh P, Lupp C, McCann J, Millikan D, Schaefer A, Stabb E, Stevens A, Visick K, Whistler C, Greenberg EP (2005) Complete genome sequence of Vibrio fischeri: A symbiotic bacterium with pathogenic congeners. Proc Natl Acad Sci USA 102:3004–3009
Schaefer AL, Val DL, Hanzelka BL, Cronan JE Jr, Greenberg EP (1996) Generation of cell-to-cell signals in quorum sensing: acyl homoserine lactone synthase activity of a purified Vibrio fischeri LuxI protein. Proc Natl Acad Sci USA 93:9505–9509
Schleicher TR, Nyholm SV (2011) Characterizing the host and symbiont proteomes in the association between the Bobtail squid, Euprymna scolopes, and the bacterium Vibrio fischeri. PloS One 6:e25649
Schuster BM, Perry LA, Cooper VS, Whistler CA (2010) Breaking the language barrier: experimental evolution of non-native Vibrio fischeri in squid tailors luminescence to the host. Symbiosis 51:85–96
Septer AN, Bose JL, Dunn AK, Stabb EV (2010) FNR-mediated regulation of bioluminescence and anaerobic respiration in the light-organ symbiont Vibrio fischeri. FEMS Microbiol Lett 306:72–81
Septer AN, Wang Y, Ruby EG, Stabb EV, Dunn AK (2011) The haem-uptake gene cluster in Vibrio fischeri is regulated by Fur and contributes to symbiotic colonization. Environ Microbiol 13(11):2855–64
Shears JS (1988) The use of a sand-coat in relation to feeding and diel activity in the sepiolid squid Euprymna scolopes. Malacologia 29:121–133
Singley CT (1983) Euprymna scolopes. In: Boyle PR (ed) Cephalopod life cycles. Academic, London, pp 69–74
Small A, McFall-Ngai M (1999) Halide peroxidase in tissues that interact with bacteria in the host squid Euprymna scolopes. J Cell Biochem 72:445–457
Soto W, Gutierrez J, Remmenga MD, Nishiguchi MK (2009) Salinity and temperature effects on physiological responses of Vibrio fischeri from diverse ecological niches. Microb Ecol 57:140–150
Spiro S (2010) An alternative route to nitric oxide resistance. Mol Microbiol 77:6–10
Stabb EV (2005) Shedding light on the bioluminescence “paradox”. ASM News 71:223–229
Stabb EV, Millikan DS (2009) Is the Vibrio fischeri-Euprymna scolopes symbiosis a defensive mutualism? In: White JF Jr, Torres MS (eds) Defensive mutualism in microbial symbiosis. Taylor and Francis, Boca Raton, pp 85–98
Stabb EV, Reich KA, Ruby EG (2001) Vibrio fischeri genes hvnA and hvnB encode secreted NAD(+)-glycohydrolases. J Bacteriol 183:309–317
Stabb EV, Ruby EG (2003) Contribution of pilA to competitive colonization of the squid Euprymna scolopes by Vibrio fischeri. Appl Environ Microbiol 69:820–826
Stabb EV, Schaefer A, Bose JL, Ruby EG (2008) Quorum signaling and symbiosis in the marine luminous bacterium Vibrio fischeri. In: Winans SC, Bassler BL (eds) Chemical communication among microbes. ASM Press, Washington, DC, pp 233–250
Stabili L, Gravili C, Tredici SM, Piraino S, Tala A, Boero F, Alifano P (2008) Epibiotic Vibrio luminous bacteria isolated from some hydrozoa and bryozoa species. Microb Ecol 56:625–636
Studer SV, Mandel MJ, Ruby EG (2008) AinS quorum sensing regulates the Vibrio fischeri acetate switch. J Bacteriol 190:5915–5923
Sugita H, Ito Y (2006) Identification of intestinal bacteria from Japanese flounder (Paralichthys olivaceus) and their ability to digest chitin. Lett Appl Microbiol 43:336–342
Sycuro L, Ruby EG, McFall-Ngai MJ (2006) Confocal microscopy of the light organ crypts in juvenile Euprymna scolopes reveals their mophological complexity and dynamic function in symbiosis. J Morphol 267:555–568
Takeuchi K, Kiefer P, Reimmann C, Keel C, Dubuis C, Rolli J, Vorholt JA, Haas D (2009) Small RNA-dependent expression of secondary metabolism is controlled by Krebs cycle function in Pseudomonas fluorescens. J Biol Chem 284:34976–34985
Tantillo GM, Fontanarosa M, Di Pinto A, Musti M (2004) Updated perspectives on emerging vibrios associated with human infections. Lett Appl Microbiol 39:117–126
Tomarev SI, Zinovieva RD, Weis VM, Chepelinsky AB, Piatigorsky J, McFall-Ngai MJ (1993) Abundant mRNAs in the squid light organ encode proteins with a high similarity to mammalian peroxidases. Gene 132:219–226
Tong D, Rozas NS, Oakley TH, Mitchell J, Colley NJ, McFall-Ngai MJ (2009) Evidence for light perception in a bioluminescent organ. Proc Natl Acad Sci USA 106:9836–9841
Troll JV, Adin DM, Wier AM, Paquette N, Silverman N, Goldman WE, Stadermann FJ, Stabb EV, McFall-Ngai MJ (2009a) Peptidoglycan induces loss of a nuclear peptidoglycan recognition protein during host tissue development in a beneficial animal-bacterial symbiosis. Cell Microbiol 11:1114–1127
Troll JV, Bent EH, Pacquette N, Wier AM, Goldman WE, Silverman N, McFall-Ngai MJ (2009b) Taming the symbiont for coexistence: a host PGRP neutralizes a bacterial symbiont toxin. Environ Microbiol 12:2190–2203
Tu S-C, Mager HIX (1995) Biochemistry of bacterial bioluminescence. Photochem Photobiol 62:615–624
Visick KL (2009) An intricate network of regulators controls biofilm formation and colonization by Vibrio fischeri. Mol Microbiol 74:782–789
Visick KL, Foster J, Doino J, McFall-Ngai M, Ruby EG (2000) Vibrio fischeri lux genes play an important role in colonization and development of the host light organ. J Bacteriol 182:4578–4586
Visick KL, O’Shea TM, Klein A, Geszvain K, Wolfe AJ (2007) The sugar phosphotransferase system of Vibrio fischeri inhibits both motility and bioluminescence. J Bacteriol 189:2571–2574
Visick KL, Ruby EG (1998) The periplasmic, group III catalase of Vibrio fischeri is required for normal symbiotic competence and is induced both by oxidative stress and by approach to stationary phase. J Bacteriol 180:2087–2092
Visick KL, Skoufos LM (2001) Two-component sensor required for normal symbiotic colonization of Euprymna scolopes by Vibrio fischeri. J Bacteriol 183:835–842
Walker EL, Bose JL, Stabb EV (2006) Photolyase confers resistance to UV light but does not contribute to the symbiotic benefit of bioluminescence in Vibrio fischeri ES114. Appl Environ Microbiol 72:6600–6606
Wang Y, Dufour YS, Carlson HK, Donohue TJ, Marletta MA, Ruby EG (2010a) H-NOX-mediated nitric oxide sensing modulates symbiotic colonization by Vibrio fischeri. Proc Natl Acad Sci USA 107:8375–8380
Wang Y, Dunn AK, Wilneff J, McFall-Ngai MJ, Spiro S, Ruby EG (2010b) Vibrio fischeri flavohaemoglobin protects against nitric oxide during initiation of the squid-Vibrio symbiosis. Mol Microbiol 78:903–915
Wei SL, Young RE (1989) Development of symbiotic bacterial bioluminescence in a nearshore cephalopod, Euprymna scolopes. Mar Biol 103:541–546
Weis VM, Small AL, McFall-Ngai MJ (1996) A peroxidase related to the mammalian antimicrobial protein myeloperoxidase in the Euprymna-Vibrio mutualism. Proc Natl Acad Sci USA 93:13683–13688
Welch RA, Burland V, Plunkett G 3rd, Redford P, Roesch P, Rasko D, Buckles EL, Liou SR, Boutin A, Hackett J, Stroud D, Mayhew GF, Rose DJ, Zhou S, Schwartz DC, Perna NT, Mobley HL, Donnenberg MS, Blattner FR (2002) Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc Natl Acad Sci USA 99:17020–17024
Whistler CA, Koropatnick TA, Pollack A, McFall-Ngai MJ, Ruby EG (2007) The GacA global regulator of Vibrio fischeri is required for normal host tissue responses that limit subsequent bacterial colonization. Cell Microbiol 9:766–778
Whistler CA, Ruby EG (2003) GacA regulates symbiotic colonization traits of Vibrio fischeri and facilitates a beneficial association with an animal host. J Bacteriol 185:7202–7212
Wier AM, Nyholm SV, Mandel MJ, Massengo-Tiasse RP, Schaefer AL, Koroleva I, Splinter-Bondurant S, Brown B, Manzella L, Snir E, Almabrazi H, Scheetz TE, Bonaldo Mde F, Casavant TL, Soares MB, Cronan JE, Reed JL, Ruby EG, McFall-Ngai MJ (2010) Transcriptional patterns in both host and bacterium underlie a daily rhythm of anatomical and metabolic change in a beneficial symbiosis. Proc Natl Acad Sci USA 107:2259–2264
Wolfe AJ, Berg HC (1989) Migration of bacteria in semisolid agar. Proc Natl Acad Sci USA 86:6973–6977
Wolfe AJ, Millikan DS, Campbell JM, Visick KL (2004) Vibrio fischeri σ54 controls motility, biofilm formation, luminescence and colonization. Appl Environ Microbiol 70:2520–2524
Wolfe AJ, Visick KL (2010) The second messenger cyclic di-GMP. ASM Press, Washington, DC
Wollenberg MS, Preheim SP, Polz MF, Ruby EG (2011) Polyphyly of non-bioluminescent Vibrio fischeri sharing a lux-locus deletion. Environ Microbiol 14(3):655–68
Wollenberg MS, Ruby EG (2009) Population structure of Vibrio fischeri within the light organs of Euprymna scolopes squid from Two Oahu (Hawaii) populations. Appl Environ Microbiol 75:193–202
Wollenberg MS, Ruby EG (2012) Phylogeny and fitness of Vibrio fischeri from the light organs of Euprymna scolopes in two Oahu, Hawaii populations. Isme J 6(2):352–62
Wright KJ, Seed PC, Hultgren SJ (2007) Development of intracellular bacterial communities of uropathogenic Escherichia coli depends on type 1 pili. Cell Microbiol 9:2230–2241
Yildiz FH, Dolganov NA, Schoolnik GK (2001) VpsR, a member of the response regulators of the two-component regulatory systems, is required for expression of vps biosynthesis genes and EPS(ETr)-associated phenotypes in Vibrio cholerae O1 El Tor. J Bacteriol 183:1716–1726
Yildiz FH, Visick KL (2009) Vibrio biofilms: so much the same yet so different. Trends Microbiol 17:109–118
Yip ES, Geszvain K, DeLoney-Marino CR, Visick KL (2006) The symbiosis regulator RscS controls the syp gene locus, biofilm formation and symbiotic aggregation by Vibrio fischeri. Mol Microbiol 62:1586–1600
Yip ES, Grublesky BT, Hussa EA, Visick KL (2005) A novel, conserved cluster of genes promotes symbiotic colonization and σ54-dependent biofilm formation by Vibrio fischeri. Mol Microbiol 57:1485–1498
Zenno S, Saigo K (1994) Identification of the genes encoding NAD(P)H-flavin oxidoreductases that are similar in sequence to Escherichia coli Fre in four species of luminous bacteria: Photorhabdus luminescens, Vibrio fischeri, Vibrio harveyi, and Vibrio orientalis. J Bacteriol 176:3544–3551
Ziegler MM, Baldwin TO (1981) Biochemistry of bacterial bioluminescence. In: Current topics in bioenergetics. Academic, Chestnut Hill, pp 65–113
Zuber S, Carruthers F, Keel C, Mattart A, Blumer C, Pessi G, Gigot-Bonnefoy C, Schnider-Keel U, Heeb S, Reimmann C, Haas D (2003) GacS sensor domains pertinent to the regulation of exoproduct formation and to the biocontrol potential of Pseudomonas fluorescens CHA0. Mol Plant Microbe Interact: MPMI 16:634–644
Acknowledgments
We thank Jonathan Visick, Andrew Weir, and Mark Mandel for reading and advising on parts of the manuscript, and members of our labs for feedback on the manuscript. We also thank individuals who contributed unpublished images, including Spencer Nyholm, Andrew Collins, Jeffrey Bose, Kati Geszvain, Satoshi Shibata, Emily Yip, and Cindy DeLoney-Marino. Work on the Vibrio fischeri-Euprymna scolopes symbiosis in the laboratories of the authors was funded by NSF grant IOS-1121106 to EVS and by NIH grant GM59690 awarded to KLV.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2013 Springer-Verlag Berlin Heidelberg
About this entry
Cite this entry
Stabb, E.V., Visick, K.L. (2013). Vibrio fisheri: Squid Symbiosis. In: Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F. (eds) The Prokaryotes. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-30194-0_22
Download citation
DOI: https://doi.org/10.1007/978-3-642-30194-0_22
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-30193-3
Online ISBN: 978-3-642-30194-0
eBook Packages: Biomedical and Life SciencesReference Module Biomedical and Life Sciences