Abstract
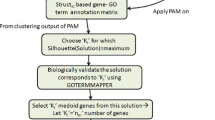
Classification of microarray cancer data has drawn the attention of research community for better clinical diagnosis in last few years. Microarray datasets are characterized by high dimension and small sample size. Hence, the conventional wrapper methods for relevant gene selection cannot be applied directly on such datasets due to large computation time. In this paper, a two stage approach is proposed to determine a subset containing relevant and non redundant genes for better classification of microarray data. In first stage, genes were partitioned into distinct clusters to identify redundant genes. To determine the better choice of clustering algorithm to group redundant genes, four different clustering methods were investigated. Experiments on four well known cancer microarray datasets depicted that hierarchical agglomerative with complete link approach performed the best in terms of average classification accuracy for three datasets. Comparison with other state-of-art methods have shown that the proposed approach which involves gene clustering is effective in reducing redundancy among selected genes to provide better classification.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
Bellman, R.: Adaptive Control Processes. In: A Guided Tour. Princeton University Press (1961)
Guyon, I., Elisseeff, A.: An Introduction to Variable and feature Selection. Journal of Machine Learning Research (3), 1157–1182 (2003)
Jain, A.K., Murthy, M.K., Flynn, P.J.: Data Clustering: A Review ACM Computing surveys, vol. 31, pp. 264–318
Rui, X., Wunsch, D.: Survey of Clustering Algorithms. IEEE Transactions on Neural Networks 16(3) (2005)
Golub, T.R., Slonim, D.K., Tamayo, P., et al.: Molecular classification of cancer: Class discovery and class prediction by gene expression monitoring. Science 286, 531–537 (1999)
Yang, K., Cai, Z., Li, J., Lin, G.H.: A stable gene selection in microarray data analysis. BMC Bioinformatics (2006) 1471-2105-7-228
Dudoit, S., Fridlyand, J., Speed, T.P.: Comparison of discrimination methods for the classification of tumors using gene expression data. Journal of the American Statistical Association 97(457), 77–87 (2002)
Kohavi, R., John, G.H.: Wrappers for feature subset selection. Artificial Intelligence 97(1-2), 273–324 (1997)
Tseng, G.C., Wong, W.H.: Tight Clustering: A Resampling-based Approach for Identifying Stable and Tight Patterns in Data Biometrics, vol. 61, pp. 10–16 (2005)
Au, W.H., Chan, K.C., Wong, A.K., Wang, Y.: Attribute Clustering for Grouping, Selection, and Classification of Gene Expression Data. IEEE/ACM Transactions on Computational Biology and Bioinformatics (TCBB) 2(2), 83–101 (2005)
Cai, Z., Xu, L., et al.: Using clustering to identify discriminatory genes with higher classification accuracy. In: IEEE Symposium 0-7695-2727-2/06
Mukhopadhyay, A., et al.: Simultaneous Informative Gene Selection and Clustering through Multiobjective Optimization. IEEE Congress on Evol. Comp., 1–8 (2010)
Tavazoie, S., Huges, D., Campbell, M.J., Cho, R.J., Church, G.M.: Systematic determination of genetic network architecture. Nature Genet., 281–285 (1999)
Eisen, M.B., Spellman, T.P., Brown, P.O., Botstein, D.: Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. USA 95(25), 14863–14868 (1998)
Pal, S.K., Mitra, P.: Pattern recognition algorithms for data mining. Chap. and Hall (2008)
Bala, R., Agrawal, R.K., Sardana, M.: Relevant Gene Selection Using Normalized Cut Clustering with Maximal Compression Similarity Measure. In: Zaki, M.J., Yu, J.X., Ravindran, B., Pudi, V. (eds.) PAKDD 2010, Part II. LNCS, vol. 6119, pp. 81–88. Springer, Heidelberg (2010)
Kohonen, T.: Self-organizing maps. Springer, Berlin (1995)
Su, A.I., Welsh, J.B.: Molecular classification of human carcinomas by gene expression signatures. Cancer Research 61, 7388–7393 (2001)
Ramaswamy, S., Tamayo, P., Rifkin, R., et al.: Multi-class cancer diagnosis using tumor gene expression signatures. PNAS 98, 15149–15154 (2001), Dataset description
Ross, D.T., Scherf, U., Eisen, M.B., Perou, C.M., et al.: Systematic Variation in Gene Expression Patterns in Human Cancer Cell Lines. Nature Genet. 24, 227–235 (2000)
Alon, U., Barkai, N., et al.: Broad Patterns of Gene Expression Revealed by Clustering Analysis of Tumor and Normal Colon Tissues Probed by Oligonucleotide Array. Proc. Nat’l Academy of Science 96(12), 6745–6750 (1999)
Zhang, Y., Ding, C., Li, T.: Gene selection algorithm by combining Relief and RMR. BMC Genomics 9(2), S27 (2008)
Wang, L., Chu, F., Xie, W.: Accurate cancer classification using expressions of very few genes. IEEE/ACM Trans. on Comp. Biology and Bioinformatics 4(1) (2007)
Cho, J., Lee, D., Park, J.H., Lee, I.B.: New gene selection for classification of cancer subtype considering within-class variation. FEBS Letters 551, 3–7 (2003)
Zhu, Z., Ong, Y.-S., Monoranjan, D.: Wrapper–Filter Feature Selection Algorithm Using a Memetic Framework. IEEE Trans. Cybernatics 37(1) (2007)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2012 Springer-Verlag GmbH Berlin Heidelberg
About this paper
Cite this paper
Sardana, M., Agrawal, R.K. (2012). A Comparative Study of Clustering Methods for Relevant Gene Selection in Microarray Data. In: Wyld, D., Zizka, J., Nagamalai, D. (eds) Advances in Computer Science, Engineering & Applications. Advances in Intelligent and Soft Computing, vol 166. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-30157-5_78
Download citation
DOI: https://doi.org/10.1007/978-3-642-30157-5_78
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-30156-8
Online ISBN: 978-3-642-30157-5
eBook Packages: EngineeringEngineering (R0)