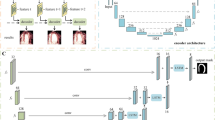
Abstract
Most existing deep learning-based frameworks for image segmentation assume that a unique ground truth is known and can be used for performance evaluation. This is true for many applications, but not all. Myocardial segmentation of Myocardial Contrast Echocardiography (MCE), a critical task in automatic myocardial perfusion analysis, is an example. Due to the low resolution and serious artifacts in MCE data, annotations from different cardiologists can vary significantly, and it is hard to tell which one is the best. In this case, how can we find a good way to evaluate segmentation performance and how do we train the neural network? In this paper, we address the first problem by proposing a new extended Dice to effectively evaluate the segmentation performance when multiple accepted ground truth is available. Then based on our proposed metric, we solve the second problem by further incorporating the new metric into a loss function that enables neural networks to flexibly learn general features of myocardium. Experiment results on our clinical MCE data set demonstrate that the neural network trained with the proposed loss function outperforms those existing ones that try to obtain a unique ground truth from multiple annotations, both quantitatively and qualitatively. Finally, our grading study shows that using extended Dice as an evaluation metric can better identify segmentation results that need manual correction compared with using Dice.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Mce dataset. https://github.com/dewenzeng/MCE_dataset
Beresford, M.J., Padhani, A.R., et al.: Inter-and intraobserver variability in the evaluation of dynamic breast cancer MRI. J. Magn. Reson. Imaging Official J. Int. Soc. Magn. Reson. Med. 24(6), 1316–1325 (2006)
Butakoff, C., Balocco, S., Ordas, S.: Simulated 3d ultrasound lv cardiac images for active shape model training. In: Medical Imaging 2007: Image Processing, vol. 6512, p. 65123U. International Society for Optics and Photonics (2007)
Chen, L.C., Zhu, Y., Papandreou, G., Schroff, F., Adam, H.: Encoder-decoder with atrous separable convolution for semantic image segmentation. In: Proceedings of the European Conference on Computer Vision (ECCV), pp. 801–818 (2018)
Dewey, M., et al.: Clinical quantitative cardiac imaging for the assessment of myocardial ischaemia. Nat. Rev. Cardiol. 17(7), 427–450 (2020)
Isensee, F., Petersen, J., et al.: nnu-net: Self-adapting framework for u-net-based medical image segmentation. arXiv preprint arXiv:1809.10486 (2018)
Litjens, G., et al.: A survey on deep learning in medical image analysis. Med. Image Anal. 42, 60–88 (2017)
Liu, Z., et al.: Machine vision guided 3d medical image compression for efficient transmission and accurate segmentation in the clouds. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 12687–12696 (2019)
McErlean, A., et al.: Intra-and interobserver variability in CT measurements in oncology. Radiology 269(2), 451–459 (2013)
Porter, T.R., Mulvagh, S.L., Abdelmoneim, S.S., Becher, H., et al.: Clinical applications of ultrasonic enhancing agents in echocardiography: 2018 American society of echocardiography guidelines update. J. Am. Soc. Echocardiogr. 31(3), 241–274 (2018)
Ronneberger, O., Fischer, P., Brox, T.: U-net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Sudre, C.H., et al.: Let’s agree to disagree: learning highly debatable multirater labelling. In: Shen, D., et al. (eds.) MICCAI 2019. LNCS, vol. 11767, pp. 665–673. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-32251-9_73
Tang, M.X., et al.: Quantitative contrast-enhanced ultrasound imaging: a review of sources of variability. Interface Focus 1(4), 520–539 (2011)
Tanno, R., Saeedi, A., Sankaranarayanan, S., Alexander, D.C., Silberman, N.: Learning from noisy labels by regularized estimation of annotator confusion. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 11244–11253 (2019)
Wang, H., Suh, J.W., Das, S.R., Pluta, J.B., Craige, C., Yushkevich, P.A.: Multi-atlas segmentation with joint label fusion. IEEE Trans. Pattern Anal. Mach. Intell. 35(3), 611–623 (2012)
Warfield, S.K., Zou, K.H., Wells, W.M.: Simultaneous truth and performance level estimation (staple): an algorithm for the validation of image segmentation. IEEE Trans. Med. Imaging 23(7), 903–921 (2004)
Xu, X., et al.: Whole heart and great vessel segmentation in congenital heart disease using deep neural networks and graph matching. In: Shen, D., et al. (eds.) MICCAI 2019. LNCS, vol. 11765, pp. 477–485. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-32245-8_53
Yaniv, Z., Lowekamp, B.C., Johnson, H.J., Beare, R.: Simpleitk image-analysis notebooks: a collaborative environment for education and reproducible research. J. Digit. Imaging 31(3), 290–303 (2018)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this paper
Cite this paper
Zeng, D. et al. (2021). Segmentation with Multiple Acceptable Annotations: A Case Study of Myocardial Segmentation in Contrast Echocardiography. In: Feragen, A., Sommer, S., Schnabel, J., Nielsen, M. (eds) Information Processing in Medical Imaging. IPMI 2021. Lecture Notes in Computer Science(), vol 12729. Springer, Cham. https://doi.org/10.1007/978-3-030-78191-0_37
Download citation
DOI: https://doi.org/10.1007/978-3-030-78191-0_37
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-78190-3
Online ISBN: 978-3-030-78191-0
eBook Packages: Computer ScienceComputer Science (R0)