Abstract
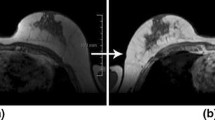
Magnetic Resonance Imaging (MRI) is currently considered to be the most sensitive tool for imaging-based diagnostic and presurgical assessment of breast cancer. In addition to their valuable diagnostic contrasts, MRI scans also provide a superb delineation of breast anatomy, which facilitates a number of related clinical applications, among which are breast density estimation and bio-mechanical modeling of breast tissue. Such applications, however, require one to know the disposition of various types of the tissue, thus warranting the procedure of image segmentation. In the case of breast MRI, the latter is known to be a challenging problem due to the relatively complicate nature of measurement noises, which is particularly problematic to deal with in the presence of bias fields. Accordingly, in this work, we introduce a simple method that can be used to “gaussianize” the noise statistic, which allows the problem of image segmentation to be formulated in the form of a simple optimization problem. In this formulation, segmentation of breast MRI scans can be completed in only a few iterations as demonstrated by our experiments involving both in silico and in vivo MRI data.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
Notes
- 1.
In practical computations, a \(3\times 3\) median filter has proven to be an adequate choice.
- 2.
In the absence of contrast enhancement, dermal and tumorous tissues have a visual appearance similar to that of dense tissue. For this reason, tumors are often included in the “dense” class.
References
Monticciolo, D.L., Newell, M.S., Moy, L., Niell, B., Monsees, B., Sickles, E.A.: Breast cancer screening in women at higher than average risk: recommendations from the ACR. J. Am. Coll. Radiol. 15(3), 408–414 (2018)
Plana, M.N., et al.: Magnetic resonance imaging in the preoperative assessment of patients with primary breast cancer: systematic review of diagnostic accuracy and meta-analysis. Eur. Radiol. 22(1), 26–38 (2012)
Song, H., Cui, X., Sun, F.: Tissue 3D segmentation and visualization on MRI. Int. J. Biomed. Imaging (2013). Article ID 859746
Fooladivanda, A., Shokouhi, S.B., Mosavi, M.R., Ahmadinejad, N.: Atlas-based automatic breast MRI segmentation using pectoral muscle and chest region model. In: ICBME (2014)
Raba, D., Oliver, A., Martí, J., Peracaula, M., Espunya, J.: Breast segmentation with pectoral muscle suppression on digital mammograms. In: Marques, J.S., Pérez de la Blanca, N., Pina, P. (eds.) IbPRIA 2005. LNCS, vol. 3523, pp. 471–478. Springer, Heidelberg (2005). https://doi.org/10.1007/11492542_58
Kannan, S.R., Ramathilagam, S., Sathya, A.: Robust fuzzy C-means in classifying breast tissue regions. In: ARTCom 2009, pp. 543–545 (2009)
Niukkanen, A., et al.: Quantitative volumetric K-means cluster segmentation of fibroglandular tissue and skin in breast MRI. J. Digit. Imaging 31(4), 425–434 (2018)
Pathmanathan, P.: Predicting tumour location by simulating the deformation of the breast using nonlinear elasticity and the finite element method. Doctoral dissertation, Wolfson College University of Oxford (2006)
Nie, K., et al.: Development of a quantitative method for analysis of breast density based on three dimensional breast MRI. Med. Phys. 35(12), 5253–5262 (2008)
Dalmıs, M.U., et al.: Using deep learning to segment breast and fibroglandular tissue in MRI volumes. Med. Phys. 44(2), 533–546 (2017)
Han, L., et al.: A nonlinear biomechanical model based registration method for aligning prone and supine MRI breast images. IEEE Trans. Med. Imaging 33(3), 682–694 (2014)
Gubern-Merida, A., Kallenberg, M., Mann, R.M., Marti, R., Karssemeijer, N.: Breast segmentation and density estimation in breast MRI: a fully automatic framework. IEEE J. Biomed. Health Inform. 19(1), 349–357 (2015)
Dempster, A.P., Laird, N.M., Rubin, D.B.: Maximum likelihood from incomplete data via the EM algorithm. J. Roy. Stat. Soc. B (Methodol.) 39(1), 1–38 (1977)
Gudbjartsson, H., Patz, S.: Rician distribution of noisy MRI data. Magn. Reson. Med. 34(6), 910–914 (1995)
Leys, C., Ley, C., Klein, O., Bernard, P., Licata, L.: Detecting outliers: do not use standard deviation around the mean, use absolute deviation around the median. J. Exp. Soc. Psychol. 49(4), 764–766 (2013)
Boyd, S., Parikh, N., Chu, E., Peleato, B., Eckstein, J.: Distributed optimization and statistical learning via the alternating direction method of multipliers. Found. Trends Mach. Learn. 3(1), 1–122 (2011)
Juntu, J., Sijbers, J., Van Dyck, D., Gielen, J.: Bias field correction for MRI images. In: Kurzyński, M., Puchała, E., Woźniak, M., Żołnierek, A. (eds.) Computer Recognition Systems. AINSC, vol. 30, pp. 543–551. Springer, Heidelberg (2005). https://doi.org/10.1007/3-540-32390-2_64
Mory, B., Ardon, R., Thiran, J.P.: Variational segmentation using fuzzy region competition and local non-parametric probability density functions. In: ICCV, pp. 1–8 (2007)
Lorentz, G.G.: Bernstein Polynomials. American Mathematical Society, Providence (2013)
Rudin, L.I., Osher, S., Fatemi, E.: Nonlinear total variation based noise removal algorithms. Physica D 60(1–4), 259–268 (1992)
Chen, D.Q., Zhang, H., Cheng, L.Z.: A fast fixed point algorithm for total variation deblurring and segmentation. J. Math. Imaging Vis. 43(3), 167–179 (2012)
Boyd, S., Vandenberghe, L.: Convex Optimization. Cambridge University Press, Cambridge (2004)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Soleimani, H., Rincon, J., Michailovich, O.V. (2019). Segmentation of Breast MRI Scans in the Presence of Bias Fields. In: Karray, F., Campilho, A., Yu, A. (eds) Image Analysis and Recognition. ICIAR 2019. Lecture Notes in Computer Science(), vol 11662. Springer, Cham. https://doi.org/10.1007/978-3-030-27202-9_34
Download citation
DOI: https://doi.org/10.1007/978-3-030-27202-9_34
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-27201-2
Online ISBN: 978-3-030-27202-9
eBook Packages: Computer ScienceComputer Science (R0)