Abstract
Delineation of organs at risk (OAR) on CT images is a crucial step in the planning of radiotherapy treatment. Manual delineation is time-consuming and high interrater variability is observed within and across radiotherapy centers. Automated delineation of OAR is fast and can lead to more consistent treatment plans. We developed an auto-delineation tool based on a 3D convolutional neural network (CNN) to automatically delineate 16 OAR structures in head and neck cancer (HNC) patients. The CNN was trained off-line using 70 previously collected patient datasets and implemented to be available on-line in clinical routine practice. The tool was applied prospectively for delineation of 20 consecutive new HNC cases within the department of Radiation Oncology, with subsequent manual editing and approval of the contours by the clinical expert. Validation based on the automatically proposed and edited contours shows that the auto-delineation tool is able to achieve highly accurate segmentation results for most OAR. As a result, 3D delineation time is reduced to less than 19 min on average (about 1 min/structure), compared to usually 1 h or more without auto-delineation tool.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Cancer is a major disease worldwide with head and neck cancers (HNC) among the most common cancers in Europe [1, 20]. State of the art treatment of patients diagnosed with HNC often involves external beam radiotherapy (RT). Treatment planning systems (TPS) are used in radiotherapy to determine an optimal treatment plan for each patient. Precise delivery of ionizing radiation to the tumor increases probability of local tumor control while maximally sparing healthy tissue in order to avoid treatment complications.
Accurate delineation of target volumes and OAR on the planning CT is required to ensure proper plan and dose optimization. In clinical practice, the delineation is performed manually by radiation oncologists (RO) based on published guidelines and is time consuming [12]. The delineation strongly depends on experience level, knowledge and preferences of a RO, leading to high intra- and interobserver variability [2]. Consequently, the induced variations may affect the final treatment plan [2, 14]. Automatic delineation can improve accuracy, consistency and reproducibility of contours leading to more consistent treatment plans within and across radiotherapy centers [12, 19].
Atlas-based models are widely used to automatically segment OAR in HNC [4, 12, 19]. Prior knowledge is incorporated in the form of atlases, which are registered to the target image using deformable image registration techniques [12].
Recently, machine learning approaches, in particular deep learning based on convolutional neural networks (CNN), proved their success in many computer vision tasks such as object detection [5], semantic segmentation [11] and classification [8, 10, 18] and are becoming a state-of-the-art approach in medical imaging as well (e.g. [7, 16]), including RT planning (e.g. [13]). For HNC in particular, Ibragimov et al. [6] developed a convolutional neural network extended with Markov random fields for segmentation of OAR in HNC patients. Men et al. [12] published a deep deconvolutional network focusing on the auto-delineation of the target volumes in HNC patients. Cardenas et al. [3] used convolutional neural networks for delineation of high risk oropharyngeal target volumes.
To investigate the clinical potential of CNN-based auto-delineation, we developed and implemented such a tool and integrated it within the conventional planning workflow within the department of Radiation Oncology of UZ Leuven. The tool is applied on-line, i.e. results are available to the radiation oncologist within few minutes after invoking the tool at the start of the planning procedure. The tool generates delineations of multiple (up to 16) organs at once, including: brainstem, spinal cord, parotid glands, submandibular glands, mandible, oral cavity, left and right cochlea, supraglottic and glottic larynx, upper esophagus and pharyngeal constrictor muscles (PCM). The auto-delineation results are imported in the planning system and visually inspected and edited as needed by the clinical expert. We report on our initial clinical experience with a quantitative and qualitative evaluation of the tool based on clinical feedback for 20 actual planning cases. Auto-delineated contours are generally well perceived by the radiation oncologists and reduce overall delineation time drastically. Due to the generic nature of the underlying CNN, the implementation is easily extendable to other organs.
2 Materials and Methods
2.1 Data Acquisition
The dataset used for training of the CNN consist of planning CT images of 70 patients and their OAR delineations. All patients were diagnosed with a tumor in head and neck region and received RT treatment. All CT images were acquired using the same clinical protocol on the same CT scanner in our institute (Somatom Sensation Open, Siemens Healthcare, Forchheim Germany). During CT acquisition, all patients were immobilized in treatment position using a thermoplastic mask. The auto-delineation tool is validated on planning CT images of 20 new HNC patients, which were consecutively acquired in clinical practice between April and May 2018. The auto-delineation tool was prospectively applied to these new cases to assess both the performance of the underlying 3D CNN and the impact on the RT planning workflow in daily routine clinical practice. Two patients received right parotidectomy, one patient left parotidectomy and four patients total laryngectomy before RT treatment, which means that the right (left) parotid resp. upper esophagus, inferior pharyngeal constrictor muscle and larynx were surgically removed and were consequently not present in the planning CT image of the patient.
2.2 3D Convolutional Neural Network: DeepVoxNet
A 3D convolutional neural network (DeepVoxNet) based on previous work from Kamnitsas et al. [7], is developed to automatically segment OAR in HNC for RT treatment planning. This end-to-end automated delineation network predicts a class label for each voxel present in a CT image [15]. CT images are normalized and resampled to a voxel size of 1\(\,\times \,\)1\(\,\times \,\)3 mm\(^3\) as a preprocessing step. Data augmentation is performed by introducing Gaussian noise and randomly flipping images. For computational efficiency, a patch based approach (19\(\,\times \,\)19\(\,\times \,\)13 voxels) is used in which multiple voxels are predicted at once. The network has four inputs (instead of two in [7]) that receive subvolumes of the image at different resolutions. Each input is followed by 10 convolutional layers and is then upsampled to the original resolution. The output of these four pathways are concatenated in the feature dimension and followed by two final convolutional layers and the classification layer. This multi-scale approach allows the network to consider both fine details in the immediate neighborhood as more coarse information in a wider environment when making a prediction. The parametric ReLU is used as activation function. Adam optimizer and dropout were used during training. As postprocessing steps, connected component analysis and smoothing are performed using MeVisLab modules (version 2.7.1).
2.3 Implementation
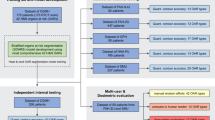
The auto-delineation tool using the proposed CNN and postprocessing steps, is deployed for testing in clinical practice within the Radiation Oncology department of UZ Leuven. New HNC planning cases follow the automated delineation protocol, which is summarized in Fig. 1. A patient’s planning CT is transferred to the Medical Image Research Center using a DICOM server (OsiriX [17]) followed by auto-delineation of the OAR using the online auto-delineation tool running on a GPU server. The auto-delineation tool is built using MeVisLab (version 2.7.1) and combines three different steps. First, preprocessing is performed by normalising the CT image and resampling it to a voxel size of 1\(\,\times \,\)1\(\,\times \,\)3 mm\(^3\). Consequently, contours of all OAR are predicted using DeepVoxNet followed by connected component analysis and smoothing as postprocessing steps. The final contours are transferred to the Radiation Oncology department in DICOM format and imported into the TPS (Eclipse, Varian Medical Systems, Palo Alto, CA, USA). If necessary the structures are corrected by a junior RO and thereafter approved by a senior RO. Corrected contours are transferred back to the Medical Image Research Center and extra clinical feedback on delineation quality and efficiency is collected. Plan and dose optimizations are performed using the standard clinical workflow. This clinical implementation allows us to gather feedback fast and efficiently to further improve auto-delineations of OAR.
2.4 Validation Process
Both a quantitative and qualitative validation is performed to assess the performance of the auto-delineation tool as well as its impact on the clinical workflow. Quantitative analysis is achieved using three similarity measures calculated in 3D including: Dice similarity coefficient (DSC), Hausdorff distance (H) and average symmetric surface distance (ASSD), which each determine the similarity between auto-delineated structures and the approved structures. Moreover, the RO recorded the time necessary to correct auto-delineated structures for each patient.
A qualitative validation is performed based on clinical assessment. The RO classifies the 3D delineation of each structure for each patient as ‘good’, ‘adequate’ or ‘insufficient’ depending on the perceived performance of the auto-delineation and the amount of extent of the manual corrections.
3 Validation Results
3.1 Quantitative Validation
Quantitative validation results reported in DSC (%), H (mm) and ASSD (mm) are summarized in Figs. 2, 3 and Table 1. The DSC values show diverse results for different anatomical structures. Brainstem, mandible, oral cavity, parotid glands, submandibular glands and spinal cord show highly accurate delineations on average, with mandible receiving the highest average DSC of 95.9% and the submandibular gland the lowest average DSC of 78.8%. Intraclass variations are rather low, which means that DeepVoxNet is able to consistently delineate the same structure. In contrast, higher intraclass variations are noticed for cochleae, pharyngeal constrictor muscles (PCM), larynx and the upper esophagus, with DSC values ranging from 0% to 100%. Cochleae are small structures and usually consist of one or two slices on the planning CT, such that even small corrections can have a large impact on DSC, resulting in a lower average DSC for the cochleae. Both the left and the right cochlea were once not recognized by the network and consequently not delineated, which explains the DSC value of 0%. The delineation results for pharyngeal constrictor muscles perform approximately the same as reported in literature [9]. Although some good auto-delineations of PCM, glottic and supraglottic larynx are obtained, leading to DSC values above 80%, the network fails to achieve accurate segmentation results when the tumor is located close to the PCM, glottic and supraglottic larynx. Moreover the transition between the PCM or glottic and supraglottic larynx are the most challenging parts to achieve high accuracy.
The average symmetric surface distance (ASSD) is below 3 mm for all structures except for the upper esophagus (7.81 mm) and the oral cavity (10.07 mm). The mandible, cochleae, spinal cord, brainstem and the right submandibular gland are the structures with the least corrections, resulting in an ASSD of less than 1 mm. Same trends are observed when evaluating Hausdorff distances. The ASSD and H highlight the influence of volume on DSC values. Although both cochleae reached lower average DSC with high intraclass variations, the cochleae achieved the best performance on ASSD scores compared to other structures.
The upper esophagus shows poor results on all three similarity measures with an average DSC of only 36.4% and Hausdorf distance of more than 3.6 cm. This can be explained by the fact that the training set only contains delineations of the upper part of the esophagus, hence labeled as ‘upper esophagus’ in Figs. 2 and 3. However, when correcting the auto-delineations, the RO extended the delineation of the upper esophagus caudally for some patients due to a lower located tumor, which explains the lower averaged similarity measures for the structure.
Figure 4 visualizes correction time per patient, recorded by the RO for correcting all the OAR delineations of a specific patient, with on average about 290 2D contours per patient. The average correction time recorded by the RO is 15 min, which is less than one minute for each 3D structure. The proposed auto-delineation tool predicts 3D contours in less than 4 min using a GPU server. This drastically decreases overall delineation time to about 19 min from approximately 45–120 min, measured in our institute.
3.2 Qualitative Validation
The auto-delineated structures are overall well perceived in clinical practice, Fig. 5. Mandible, brainstem, cochleae and spinal cord are perceived as ‘good’ for more than 80% of the cases, which is in line with the results from the quantitative validation. Every organ is more classified as ‘good’ than as ‘insufficient’ except for the upper pharyngeal constrictor muscle. The delineation of this structure needed in general more corrections, which is also observed in the quantitative validation. The upper esophagus however, scored remarkably well on the clinical score although the quantitative results are rather poor. Although the esophagus was not fully delineated, the auto-delineation of the upper part of the structure was well perceived in clinical practice.
4 Discussion and Conclusion
We developed an online auto-delineation tool for organs at risk in HNC patients in the context of RT treatment planning. The auto-delineation tool, based on 3D CNN (DeepVoxNet) is deployed in clinical practice to evaluate the performance of auto-delineations and to asses the impact on the clinical workflow.
Manual delineations are sensitive to interrater variability, leading to inconsistent treatment plans. Dice similarity coefficients of interrater variability published in literature are summarized in Table 1 [14, 19]. A high interrater variability is observed for smaller organs such as: cochleae, upper esophagus, supraglottic larynx and PCM. The inter observer variability stresses the difficulty of automatic delineations of the OAR. Segmentation results of organs at risk using both atlas-based methods and deep learning, have been reported in literature [4, 19] DeepVoxNet [15] is able to provide better segmentation results for organs at risk in head and neck patients compared to results published in literature (Table 1). Ibragimov et al. [6] was the first to propose a convolutional neural network for auto-delineations of OAR in HNC patients. Our results reported in DSC, tend to exceed the results of [6].
Our initial experience shows that in general, only small corrections are necessary for clinical acceptance of auto-delineated contours for most of the structures. The largest corrections for clinical acceptance are observed for the upper esophagus and glottic area while mandible needed the least corrections. Moreover the automated workflow is less time consuming, reducing the delineation time to 19 min in total compared to 45 min–120 min if manually delineated.
References
Borras, J.M., et al.: How many new cancer patients in Europe will require radiotherapy by 2025? An ESTRO-HERO analysis. Radiother. Oncol. 119, 5–11 (2016)
Brouwer, C.L., Steenbakkers, R.J.H.M., Heuvel, E.V.D., Duppen, J.C., Navran, A.: 3D variation in delineation of head and neck organs at risk. Radiat. Oncol. 7–32 (2012)
Cardenas, C.E., et al.: Deep learning algorithm for auto-delineation of high-risk oropharyngeal clinical target volumes with built-in dice similarity coefficient parameter optimization function. Int. J. Radiat. Oncol. Biol. Phys. 101(2), 468–478 (2018)
Fortunati, V., et al.: Tissue segmentation of head and neck CT images for treatment planning: a multiatlas approach combined with intensity modeling. Med. Phys. 40(7), 071905 (2013)
Girshick, R., Donahue, J., Darrell, T., Malik, J.: Rich feature hierarchies for accurate object detection and semantic segmentation. In: Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, pp. 580–587, June 2014
Ibragimov, B., Xing, L.: Segmentation of organs-at-risks in head and neck CT images using convolutional neural networks. Med. Phys. 44(2), 547–557 (2017)
Kamnitsas, K., et al.: Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med. Image Anal. 36, 61–78 (2017)
Krizhevsky, A., Sulskever, I., Hinton, G.E.: ImageNet classification with deep convolutional neural networks. In: Advances in Neural Information and Processing Systems (NIPS), vol. 60(6), pp. 84–90 (2012)
La Macchia, M., et al.: Systematic evaluation of three different commercial software solutions for automatic segmentation for adaptive therapy in head-and-neck, prostate and pleural cancer. Radiat. Oncol. 7(1), 1 (2012)
Litjens, G., et al.: A survey on deep learning in medical image analysis. Med. Image Anal. 42, 60–88 (2017)
Long, J., Shelhamer, E., Darrell, T.: Fully convolutional networks for semantic segmentation. In: Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, 7–12 June 2015, pp. 3431–3440 (2015)
Men, K., et al.: Deep deconvolutional neural network for target segmentation of nasopharyngeal cancer in planning computed tomography images. Front. Oncol. 7, 315 (2017)
Men, K., Dai, J., Li, Y.: Automatic segmentation of the clinical target volume and organs at risk in the planning CT for rectal cancer using deep dilated convolutional neural networks. Med. Phys. 44(12), 6377–6389 (2017)
Nelms, B.E., Tomé, W.A., Robinson, G., Wheeler, J.: Variations in the contouring of organs at risk: test case from a patient with oropharyngeal cancer. Int. J. Radiat. Oncol. Biol. Phys. 82(1), 368–378 (2012)
Robben, D., Bertels, J., Willems, S., Vandermeulen, D., Maes, F., Suetens, P.: DeepVoxNet: voxel-wise prediction of 3D images. Technical report, KU Leuven/ESAT/PSI, 1801, June 2018
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Rosset, A., Spadola, L., Ratib, O.: OsiriX: an open-source software for navigating in multidimensional DICOM images. J. Digit. Imaging 17(3), 205–216 (2004)
Sermanet, P., Eigen, D., Zhang, X., Mathieu, M., Fergus, R., Lecun, Y.: OverFeat: integrated recognition, localization and detection using convolutional networks (2014)
Tao, C.J., et al.: Multi-subject atlas-based auto-segmentation reduces interobserver variation and improves dosimetric parameter consistency for organs at risk in nasopharyngeal carcinoma: a multi-institution clinical study. Radiother. Oncol. 115(3), 407–411 (2015)
Torre, L., Siegel, R., Ward, E., Jemal, A.: Global cancer incidence and mortality rates and trends - an update. Cancer Epidemiol. Biomark. Prev. 25(1), 16–27 (2016)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this paper
Cite this paper
Willems, S. et al. (2018). Clinical Implementation of DeepVoxNet for Auto-Delineation of Organs at Risk in Head and Neck Cancer Patients in Radiotherapy. In: Stoyanov, D., et al. OR 2.0 Context-Aware Operating Theaters, Computer Assisted Robotic Endoscopy, Clinical Image-Based Procedures, and Skin Image Analysis. CARE CLIP OR 2.0 ISIC 2018 2018 2018 2018. Lecture Notes in Computer Science(), vol 11041. Springer, Cham. https://doi.org/10.1007/978-3-030-01201-4_24
Download citation
DOI: https://doi.org/10.1007/978-3-030-01201-4_24
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-01200-7
Online ISBN: 978-3-030-01201-4
eBook Packages: Computer ScienceComputer Science (R0)