Abstract
In this paper, we extend the U-Net model and propose a novel hybrid pyramid U-Net (HPU-Net) model which explores the global context information combined different region based context. Global context information combination is effective for producing good quality results in tumor segmentation tasks, and HPU-Net provides a better framework for pixel-level prediction. Because of the continuous downsampling of FCN the resolution of the feature map gradually decreases and direct upsampling during restoration of resolution will introduce noise and make the segmentation inaccurate. A novel and efficient multimodal tumor segmentation (including internal tumor) model based on U-Net is proposed to perform end-to-end training and testing. Our model includes a downsampling path and a symmetrical upsampling path, concatenating the features at the symmetrical block of upsampling and downsampling path. In the process of upsampling, we extract multiple scale features from every block, and add them pixel-wise after recovering them to origin resolution. Integrating the multi-scale information, semantic and location information before softmax layer, it helps the model complete the segmentation efficiently. The model was evaluated on two datasets BRATS2015 and BRATS2017, and outperformed state-of-the-art methods with better segmentation results.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
For clinical applications, accurate segmentation of tumors is very meaningful for diagnosis and surgical treatment. Segmentation of brain tumors from multimodal magnetic resonance imaging (MRI) is a challenging task due to the segmentation of brain tumor plays an important role in the computer-aided brain tumor disease diagnosis, while the standard for the segmentation of brain tumor is not very clear. Also the brain tumor’s various shapes and the intensity level similarity between tumor tissue and neighboring organs will result in the segmentation performance degraded. Therefore, how to accurately and efficiently segment the brain tumor, becomes a hot topic in the medical image analysis field.
There are kinds of methods to segment the brain tumor, such as the level-set, the region growing and the fuzzy clustering. Manual intervention is required in some methods. For example, the region growing method [12] needs users to manually select a seed point in the image. Recently, automatically selecting the seed point [4] has been proposed. Level-set is another segmenting method which is based on the active contour model. How to select a better initial contour is essential in the level-set. R. Rana employed the fast Bounding Algorithm to select the initial contour in the tumor area, and used the level set method to extract the tumor boundary accurately [17]. Fuzzy clustering method is usually adopted by combining with other methods, such as K-means or C-means [20]. These methods also need a prior knowledge of the data distribution. And another method is to classify the voxels into different tissues by using hand-crafted features, and then use the conditional random field (CRF) model to combine the smoothness of the classification results and maximize the consistency of the levels between the pixels in the neighborhood [15, 23].
Recently, convolutional neural networks(CNN) have achieved breakthrough achievements in various visual fields such as image classification [11], object detection [6] and natural image semantic segmentation [5, 14]. Moreover, CNN is gradually applied to brain tumor segmentation and has achieved good results. One of a popular method is to extract the image patches from the MRI through the sliding window and assign labels for the central pixel [16, 19]. As stated in [19] the image patches can be employed to complete the segmentation, adopting different cascading modes so that the model can simultaneously extract local and global information. All these methods are patch-level-based, but these methods need too much training data and time-consuming.
Fully convolutional model(FCN) [14], achieved good performance in natural image segmentation. It replaces the fully connected layers of the traditional CNN with convolutional kernels and adds upsampling to restore the resolution of the input image. FCNN [2] and DUNet [9] use a fully convolutional approach to achieve an end-to-end segmentation model. These models are all similar to U-Net model [18], but each block is internally different. In this paper, we propose a hybrid pyramid U-Net(HPU-Net) model for brain tumor segmentation. Our main contributions are as follows:
-
Feature pyramid is introduced into the U-Net model. Combining multiple scales of information to complete the segmentation.
-
Hybriding multi-scale information with the semantc and location information, improves the segmentation performance.
2 Methodology
In this section, we present a hybrid pyramid U-Net (HPU-Net) model for brain tumor segmentation. The proposed network is used to process multimodal MRI and combine multi-scale information from different stages for efficient and accurate image segmentation.
2.1 HPU-Net Model
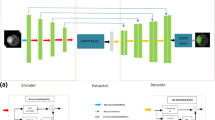
The architecture of the model is shown in Fig. 1. It consists of 3 modules, a downsampling path with convolution and max pooling layers, an upsampling path with convolution and upsample, and an auxiliary segmentation path based on the image pyramid. The downsampling path is mainly to extract high level and global contextual features of the tumor. However, the upsampling path is used to reconstruct the object details. As we know, the high-level feature has much semantic information and low-level feature has much location information, the auxiliary path is used to extract multi-scale information and make full use of multiple levels of information and combine semantic and location information in the upsampling path to help the model complete segmentation for objects of different scales.
The downsampling path is similar to U-Net’s model, but there is a slightly different. We add batch normalization (BN) [8] layer inside each block, and each block has two convolutional layers with \(3 \times 3\) kernels and two BN layers and 1 max-pooling layer with \(2 \times 2\) strides. There two main reasons for why we make these changes: (1) As the model going deeper, gradient vanishing may occur during the back-propagation which making the training of the model stagnant, and in order to speed up the convergence of the model, we add the BN layer. (2) In medical images, some lesions occupy a smaller proportion of the entire image, and as the network going deeper, convolution and each downsampling operation may cause the lesion area to vanish. So in order to extract high-level information, we use two convolutional layers in each block.
For the upsampling path, we use symmetric structures with downsampling. Each block contains two convolutional layers with \( 3 \times 3 \) kernels and two BN layers, and one upsampling layer. The feature map after upsampling, and then concatenates with the feature map before maxpooling of the symmetric block in the downsampling path, this can combine the semantic and location information. Note that we used bilinear interpolation to perform upsampling, did not use convtranspose or deconvolutional layer as it will introduce more parameters and calculations in the network. After the concatenation, the new feature map contains semantic and location information and we can obtain the better results.
2.2 Hybrid Pyramid Network
Whether in object detection or image segmentation, the network depth and stride are usually a pair of contradictory things. The commonly used network structure corresponds to a relatively large stride, and the small objects in the image are even smaller than the stride size. The segmentation performance will decrease for some small objects. Common idea for solving this problem is multi-scale training and testing, also known as image pyramids [1]. However, this approach requires high time and computational cost. In object detection, Tsung-Yi Lin [13] proposed a feature pyramid method to detect small targets. So in our proposed method, feature pyramid is proposed to integrate the multi-scale information with the semantic and location information. Figure 1 illustrates the layout of the performance HPU-Net schematically.
In the upsampling path, if we only upsample the feature map one by one block, the segmentation results will have some holes, especially for smaller tumor regions which the model may ignore. This will greatly degrade the segmentation performance. And the tumor has multi-scale shapes and size, so we employ the feature pyramid to effectively explore the multi-scale information of the objects. Then, we upsample the feature map from each block in the upsampling path to obtain the same size feature map as the original input image by bilinear interpolation. After upsampling, the feature map is then merged with the corresponding bottom-up map in the upsampling path by element-wise addition. And then a \(1 \times 1\) convolutiuonal layer is attached to reduce the channel dimensions. At last a softmax layer is applied to finish the final classification. So the softmax layer receives the output feature maps from all processing blocks in the upsampling path \(x_{0}\), \(x_{1}, ..., x_{l-1}\) as inputs:
where \(x_{i}\) means the feature maps of every block in the upsampling path, the input feature maps of softmax layer is \(X_{\text {in}\_\text {softmax}}\), H(x) refers to the operation of upsamping and convolution. The feature map used in the final prediction combines features of different scales and different semantic intensities. This not only uses multi-scale information, but also employs the semantic information in the downsampling path and the location information in the upsampling path to achieve the best segmentation results. This approach only adds 4 convolutional layers compared with U-Net, introducing a small number of parameters, but the segmentation performance has been improved significantly.
In each block of network, we use the combination of CONV-BN-RELU. In order to ensure non-linear mapping we use RELU [11] as activation function and batch normalization to reduce the internal-covariate-shift. With the BN layer, we can increase the learning rate to accelerate the convergence speed of the model and prevent the gradient vanish.
3 Evaluation
The BRATS2015 [10, 15] and BRATS2017 [3, 15] challenge dataset are used to train and validate in our experiment. The BRATS2015 training data set includes 290 samples, 220 from the high grade glioma category (HGG) and 70 from the low grade glioma (LGG) category. And BRATS2017 training dataset consisted of 210 samples from HGG and 75 samples from LGG.
Every subject has multimodal MRI: namely T1, T1-contrast (T1c), T2 and Flair, which are skull-stripped and co-registered. Figure 2 shows the gliomas tumor with doctors delineation and the internal region. The evaluation for segmentation results mainly consists of three parts: (1) complete tumor region; (2)the core region of the tumor (including all tumor area except for edema); (3) the enhancing tumor region (only including the enhancing tumor area). For each part, the Dice Similarity Coefficient (DSC), Positive Predictive Value (PPV) and Sensitivity are computed. The DSC calculates the overlap part between the manual and the automatic segmentation. It is defined as,
where FN, FP and TP are the numbers of false negative, false positive and true positive detections, respectively. Sensitivity is useful to evaluate the number of TP and FN detections, defined as,
Finally, PPV is a measure of the amount of TP and FP, defined as,
3.1 Implementation
We normalized each subject’s data with zero mean and unit standard deviation. Then we removed the slices that do not contain tumor information. And all images are cropped to 160*160 as the input to the model. At the end, BRATS2015 dataset only retained 15,000 slices, and BRATS2017 dataset retained 17800 slices. We augmented the dataset by left rotating the first half and right rotating the other half to construct a new dataset that is two times larger than the original one.
We use the Keras library with Tensorflow as the backend. The model was trained with standard back-propagation using Adam as an optimizer, and all parameters are initialized using he_normal. The training time on the augmented data is about ten hours to run 70 rounds using a standard computer with a NVIDIA Titan X GPU.
3.2 Cross Validation
We performed a 5-fold cross-validation on the augmented data and two experiments were achieved to evaluate the deeper model and hybrid pyramid.
First, we tested with four blocks and three blocks in down-sampling path, to verify whether the deeper of the model could improve the segmentation accuracy. We plotted the dice coefficients for the three tasks in different epochs. As shown in Fig. 3, it can be seen that the four block models significantly improve the partitioned dice coefficients on the three tasks compared with the model with three blocks. This is because increasing the depth of the model helps to extract more high-level features, and going deeper of the model will also provide the pyramid module with more multi-scale information. Especially for the core and enhancing tumor regions, the dice coefficients improved at least 7%. Because the area of these two regions is relatively small and the deepening of the model will integrate more multi-scale information.
We also explored the impact of the hybrid pyramid on model accuracy. Figure 4 shows the effect of dice coefficients on models with and without pyramids on the validation set. For each task, it is clear that the introduction of the pyramid improve the segmentation performance of the classification model. Without using hybrid pyramid network, the classification model degraded on the segmentation of core and enhancing tumor regions. In our experiment, the dice coefficients with hybrid pyramid network can improve 5% at least. It confirmed the improvement of our proposed model with the feature pyramid module.
3.3 Results Analysis
We compared the proposed method with state-of-the-art methods on BRATS2017 dataset. As it contains HGG and LGG images, we use the 3560 slices as test which are not involved in training. The proposed method is among the top-ranking in the state-of-the-art (see Table 1).
Specifically, FCNN and DUNet achieved good performance on BRATS2017 challenge. The performence of our model is better (by a big margin over FCNN and DUNet, e.g., 0.80 vs 0.67 and 0.80 vs 0.70 in terms of Dice for Core tumor segmentation). Particularly, FCDenseNet [21], as we know the DenseNet [7] got the best performance on ILSVRC2017. FCDenseNet references the dense block, and it’s dice and sensitivity on enhancing region is lower than HPU-Net (0.59 vs 0.76 and 0.59 vs 0.67) and the FCDenseNet needs more memory and the training time is longer than our method.
To confirm that the performance of our model, we also evaluate our proposed method BRATS2015 dataset. For the same test data, the performance of the baseline system and our proposed method on BRATS2015 is shown in Table 2. From these experimental result, we can see that our model also shows state-of-the-art performance on this dataset. Our HPU-Net network structure is simple and effective combining multi-scale features.
As we can see from Fig. 5, the segmentation results of groundtruth, DUNet, FCNN, FCDenseNet, VGG, and our proposed HPU-Net model are shown from left to right. It is clear that DUNet divided some of the necrosis regions (blue) into non-enhancing regions (orange). The FCNN directly ignored non-enhancing regions (orange). However, FCDenseNet divided some of the enhancing regions (dark red) into edema regions (green). VGGNet divided some enhancing regions (dark red) into non-enhancing regions (orange). These wrong segmentation results were due to lost of the multi-scale information of the data. On the contrary, the HPU-Net model performed better because of the effective fusion of multi-scale features.
4 Conclusion
We propose a hybrid pyramid U-Net model which is an end-to-end brain tumor segmentation model. Our model includes a downsampling path and an upsampling path and a hybrid pyramid path to extract multi-scale information. Deeper model made the dice improved, and the introduction of the feature pyramid also improved the segmentation result. Our model achieved significant better results and we try to perform the nature image segmentation in the future.
References
Adelson, E.H., Anderson, C.H., Bergen, J.R., Burt, P.J., Ogden, J.M.: Pyramid methods in image processing. RCA Eng. 29(6), 33–41 (1984)
Alex, V., Safwan, M., Krishnamurthi, G.: Brain tumor segmentation from multi modal MR images using fully convolutional neural network. In: Medical Image Computing and Computer Assisted Intervention - MICCAI 2017–20th International Conference Proceedings, pp. 1–8. Springer (2017)
Bakas, S., Akbari, H., Sotiras, A., Bilello, M., Rozycki, M., Kirby, J.S., Freymann, J.B., Farahani, K., Davatzikos, C.: Advancing the cancer genome atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci. Data 4, 170117 (2017)
Balasubramanian, C., Saravanan, S., Srinivasagan, K., Duraiswamy, K.: Automatic segmentation of brain tumor from MR image using region growing technique. Life Sci. J. 10(2), 2878–2883 (2013)
Chen, L.C., Papandreou, G., Kokkinos, I., Murphy, K., Yuille, A.L.: Deeplab: semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected CRFs. IEEE Trans. Pattern Anal. Mach. Intell. 40(4), 834–848 (2018)
Girshick, R., Donahue, J., Darrell, T., Malik, J.: Rich feature hierarchies for accurate object detection and semantic segmentation. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 580–587 (2014)
Huang, G., Liu, Z., Weinberger, K.Q., van der Maaten, L.: Densely connected convolutional networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, vol. 1, p. 3 (2017)
Ioffe, S., Szegedy, C.: Batch normalization: accelerating deep network training by reducing internal covariate shift (2015). arXiv preprint arXiv:1502.03167
Kim, G.: Brain tumor segmentation using deep u-net. In: Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D.L., Duchesne, S. (eds.) MICCAI 2017, Part III. LNCS, vol. 10435. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-66179-7
Kistler, M., Bonaretti, S., Pfahrer, M., Niklaus, R., Büchler, P.: The virtual skeleton database: an open access repository for biomedical research and collaboration. J. Med. Internet Res. 15(11), e245 (2013). https://doi.org/10.2196/jmir.2930. http://www.jmir.org/2013/11/e245/
Krizhevsky, A., Sutskever, I., Hinton, G.E.: Imagenet classification with deep convolutional neural networks. In: Advances in Neural Information Processing Systems, pp. 1097–1105 (2012)
Lenvine, M., Shaheen, S.: A modular computer vision system for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 3(5), 540–557 (1981)
Lin, T.Y., Dollár, P., Girshick, R., He, K., Hariharan, B., Belongie, S.: Feature pyramid networks for object detection. In: CVPR, vol. 1, p. 4 (2017)
Long, J., Shelhamer, E., Darrell, T.: Fully convolutional networks for semantic segmentation. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 3431–3440 (2015)
Menze, B.H., Jakab, A., Bauer, S., Kalpathy-Cramer, J., Farahani, K., Kirby, J., Burren, Y., Porz, N., Slotboom, J., Wiest, R.: The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans. Med. Imag. 34(10), 1993–2024 (2015)
Pereira, S., Pinto, A., Alves, V., Silva, C.A.: Brain tumor segmentation using convolutional neural networks in MRI images. IEEE Trans. Med. Imag. 35(5), 1240–1251 (2016)
Rana, R., Bhdauria, H., Singh, A.: Brain tumour extraction from MRI images using bounding-box with level set method. In: 2013 Sixth International Conference on Contemporary Computing (IC3), pp. 319–324. IEEE (2013)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Saha, R., Phophalia, A., Mitra, S.K.: Brain tumor segmentation from multimodal MR images using rough sets. In: Mukherjee, S., Mukherjee, S., Mukherjee, D.P., Sivaswamy, J., Awate, S., Setlur, S., Namboodiri, A.M., Chaudhury, S. (eds.) ICVGIP 2016. LNCS, vol. 10481, pp. 133–144. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-68124-5_12
Selvakumar, J., Lakshmi, A., Arivoli, T.: Brain tumor segmentation and its area calculation in brain MR images using k-mean clustering and fuzzy c-mean algorithm. In: 2012 International Conference on Advances in Engineering, Science and Management (ICAESM), pp. 186–190. IEEE (2012)
Shaikh, M., Anand, G., Acharya, G., Amrutkar, A., Alex, V., Krishnamurthi, G.: Brain tumor segmentation using dense fully convolutional neural network. In: Crimi, A., Bakas, S., Kuijf, H., Menze, B., Reyes, M. (eds.) BrainLes 2017. LNCS, vol. 10670, pp. 309–319. Springer, Cham (2018). https://doi.org/10.1007/978-3-319-75238-9_27
Shen, H., Zhang, J., Zheng, W.: Efficient symmetry-driven fully convolutional network for multimodal brain tumor segmentation. ICIP (2017, to appear) Google Scholar (2017)
Soltaninejad, M., Zhang, L., Lambrou, T., Allinson, N., Ye, X.: Multimodal mri brain tumor segmentation using random forests with features learned from fully convolutional neural network (2017). arXiv preprint arXiv:1704.08134
Acknowledgement
This work is supported by the Shandong Province Key Innovation Project (Grant no. 2017CXGC1504), the Fundamental Research Funds of Shandong University (Grant no. 2017JC013), Shandong Provincial Science and Technology Major Project (Emerging Industry) (Grant no. 2015ZDXX0801A01).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 IFIP International Federation for Information Processing
About this paper
Cite this paper
Kong, X., Sun, G., Wu, Q., Liu, J., Lin, F. (2018). Hybrid Pyramid U-Net Model for Brain Tumor Segmentation. In: Shi, Z., Mercier-Laurent, E., Li, J. (eds) Intelligent Information Processing IX. IIP 2018. IFIP Advances in Information and Communication Technology, vol 538. Springer, Cham. https://doi.org/10.1007/978-3-030-00828-4_35
Download citation
DOI: https://doi.org/10.1007/978-3-030-00828-4_35
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-00827-7
Online ISBN: 978-3-030-00828-4
eBook Packages: Computer ScienceComputer Science (R0)