Abstract
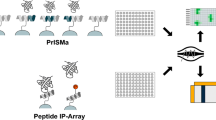
Protein–protein interactions are key elements in the assembly of cellular regulatory and signaling protein complexes that integrate and transmit signals and information in controlling and regulating various cellular processes and functions. Many conventional methods of studying protein–protein interaction, such as the immuno-precipitation and immuno-blotting assay and the affinity-column pull-down and chromatographic analysis, are very time-consuming and labor intensive and lack accuracy and sensitivity. We have developed a simple, rapid, and sensitive assay using a ProteinChip array and SELDI-TOF mass spectrometry to analyze protein–protein interactions and map the crucial elements that are directly involved in these interactions. First, a purified “bait” protein or a synthetic peptide of interest is immobilized onto the preactivated surface of a PS10 or PS20 ProteinChip and the unoccupied surfaces on the chip are protected by application of a layer ethanolamine to prevent them from binding to other non-interactive proteins. Then, the target-containing cellular protein lysate or synthetic peptide containing the predicted amino acid sequence of protein-interaction motif is applied to the protected array with immobilized bait protein/peptide. The nonspecific proteins/peptides are washed off under various stringent conditions and only the proteins specifically interacting with the bait protein/peptide remain on the chip. Last, the captured interacting protein/peptide complexes are then analyzed by SELDI-TOF mass spectrometry and their identities are confirmed by their predicted distinctive masses. This method can be used to unambiguously detect the specific protein–protein interaction of known proteins/peptides, to easily identify potential cellular targets of proteins of interest, and to accurately analyze and map the structural elements of a given protein and its target proteins using synthetic peptides with the predicted potential protein interaction motifs.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Yaffe, M. B. and Cantley, L. C. (1999) Signal transduction. Grabbing phosphoproteins. Nature. 402, 30–31.
Yaffe, M. B. and Smerdon, S. J. (2001) PhosphoSerine/threonine binding domains: you can’t pSERious? Structure. 9, R33-R38.
Schlessinger, J. (2002) A solid base for assaying protein kinase activity. Nat. Biotechnol. 20, 232–233.
Schlessinger, J. and Lemmon, M. A. (2003) SH2 and PTB domains in tyrosine kinase signaling. Sci. STKE. 2003, RE12.
Pawson, T. and Nash, P. (2000) Protein-protein interactions define specificity in signal transduction. Genes Develop. 14, 1027–1047.
Pawson, T. and Nash, P. (2003) Assembly of cell regulatory systems through protein interaction domains. Science. 300, 445–452.
Fanning, A. S. and Anderson, J. M. (1999) Protein modules as organizers of membrane structure. Curr. Opin. Cell Biol. 11, 432–439.
Fanning, A. S. and Anderson, J. M. (1998) PDZ domains and the formation of protein networks at the plasma membrane. Curr. Topics Microbiol. Immunol. 228, 209–233.
Fanning, A. S. and Anderson, J. M. (1999) Protein modules as organizers of membrane structure. Curr. Opin. Cell Biol. 11, 432–439.
Rigaut, G., Shevchenko, A., Rutz, B., Wilm, M., Mann, M., Séraphin B. (1999) A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17, 1030–1032.
Wilkins, M. R., Gasteiger, E., Gooley, A. A., Herbert, B. R., Molloy, M. P., Binz, P. A., Ou, K., Sanchez, J. C., Bairoch, A., Williams, K. L., and Hochstrasser, D. F. (1999) High-throughput mass spectrometric discovery of protein post-translational modifications. J. Mol. Biol. 289, 645–657.
Jonsson, P. F., Cavanna, T., Zicha, D., and Bates, P. A. (2006) Cluster analysis of networks generated through homology: automatic identification of important protein communities involved in cancer metastasis. BMC Bioinformatics. 7, 2–14.
Liu, Y., Porta, A., Peng, X., Gengaro, K., Cunningham, E. B., Li, H., Dominguez, L. A., Bellido, T., and Christakos, S. (2004) Prevention of glucocorticoid-induced apoptosis in osteocytes and osteoblasts by calbindin-D-28k. J. Bone Miner. Res. 19, 479–490.
Ito, I., Ji, L., Tanaka, F., Saito, Y., Gopalan, B., Branch, C. D., Xu, K., Atkinson, E. N., Bekele, B. N., Stephens, L. C., Minna, J. D., Roth, J. A., and Ramesh, R. (2004) Liposomal vector mediated delivery of the 3p FUS1 gene demonstrates potent antitumor activity against human lung cancer in vivo. Cancer Gene Ther. 11, 733–739.
Kondo, M., Ji, L., Kamibayashi, C., Tomizawa, Y., Randle, D., Sekido, Y., Yokota, J., Kashuba, V., Zabarovsky, E., Kuzmin, I., Lerman, M., Roth, J., Minna, J. D. (2001) Overexpression of candidate tumor suppressor gene FUS1 isolated from the 3p21.3 homozygous deletion region leads to G1 arrest and growth inhibition of lung cancer cells. Oncogene. 20, 6258–6262.
Liu, X., Miller, C. W., Koeffler, P. H., and Berk, A. J. (1992) The p53 activation domain binds the TATA box-binding polypeptide in Holo-TFII-D, and a neighboring p53 domain inhibits transcription. Cell. 13, 3291–3300.
Acknowledgments
This work was partially supported by grants from NIH/NCI SPORE P50CA070907, RO1CA116322, DOD PROSPECT W81XWH-0710306; and the M. D. Anderson Cancer Center Support Core Grant (CA16672) for using the Peptide Synthesis Facility to synthesize all the peptides used in this study.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2012 Springer Science+Business Media, LLC
About this protocol
Cite this protocol
Jayachandran, G., Roth, J.A., Ji, L. (2012). Analysis of Protein–Protein Interaction Using ProteinChip Array-Based SELDI-TOF Mass Spectrometry. In: Clarke, C., McCarthy, D. (eds) SELDI-TOF Mass Spectrometry. Methods in Molecular Biology, vol 818. Springer, New York, NY. https://doi.org/10.1007/978-1-61779-418-6_15
Download citation
DOI: https://doi.org/10.1007/978-1-61779-418-6_15
Published:
Publisher Name: Springer, New York, NY
Print ISBN: 978-1-61779-417-9
Online ISBN: 978-1-61779-418-6
eBook Packages: Springer Protocols