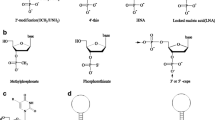
Abstract
Aptamers are nucleic acid molecules that bind to a target molecule with high affinity and specificity, which are generated by a process known as systematic evolution of ligands by exponential enrichment (SELEX). Because of their high affinity and specificity, aptamers were developed as therapeutic agents. Although aptamers are investigated as promising therapeutic agents, the mechanism of their high affinity and specificity is not clear. Therefore, structural and biophysical studies are important to know that. To date, ITC is increasingly being used to study the thermodynamic basis of aptamer–target protein interactions. Understanding the mechanism of aptamer binding would contribute to their development for therapeutic applications. In this chapter, we describe the protocol to study the thermodynamics of aptamer–protein interactions.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Tuerk C, Gold L (1990) Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249:505–510
Ellington AD, Szostak JW (1990) In vitro selection of RNA molecules that bind specific ligands. Nature 346:818–822
Stoltenburg R, Reinemann C, Strehlitz B (2007) SELEX-A (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol Eng 24:381–403
Keefe AD, Pai S, Ellington AD (2010) Aptamers as therapeutics. Nat Rev Drug Discov 9:537–548
Nakamura Y, Ishiguro A, Miyakawa S (2012) RNA plasticity and selectivity applicable to therapeutics and novel biosensor development. Genes Cells 17:344–364
Ng EW, Shima DT, Calias P, Cunningham ET Jr, Guyer DR, Adamis AP (2006) Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat Rev Drug Discov 5:123–132
Zhou B, Wang B (2006) Pegaptanib for the treatment of age-related macular degeneration. Exp Eye Res 83:615–619
Potty AS, Kourentzi K, Fang H, Jackson GW, Zhang X, Legge GB, Willson RC (2009) Biophysical characterization of DNA aptamer interactions with vascular endothelial growth factor. Biopolymers 91:145–156
Potty AS, Kourentzi K, Fang H, Schuck P, Willson RC (2011) Biophysical characterization of DNA and RNA aptamer interactions with hen egg lysozyme. Int J Biol Macromol 48:392–397
Kanakaraj I, Chen W-H, Poongavanam M, Dhamane S, Stagg LJ, Ladbury LE, Kourentzi K, Strych U, Willson RC (2013) Biophysical characterization of VEGF–aHt DNA aptamer interactions. Int J Biol Macromol 57:69–75
Cheung YW, Kwok J, Law AW, Watt RM, Kotaka M, Tanner JA (2013) Structural basis for discriminatory recognition of Plasmodium lactate dehydrogenase by a DNA aptamer. Proc Natl Acad Sci U S A 110:15967–15972
Amano R, Takada K, Tanaka Y, Nakamura Y, Kawai G, Kozu T, Sakamoto T (2016) Kinetic and Thermodynamic Analyses of Interaction between a High-Affinity RNA Aptamer and Its Target Protein. Biochemistry 55:6221–6229
Pica A, Russo Krauss I, Parente V, Tateishi-Karimata H, Nagatoishi S, Tsumoto K, Sugimoto N, Sica F (2017) Through-bond effects in the ternary complexes of thrombin sandwiched by two DNA aptamers. Nucleic Acids Res 45:461–469
Sakamoto T, Ennifar E, Nakamura Y (2017) Thermodynamic study of aptamers binding to their target proteins. Biochimie 145:91–97
Prabhu NV, Sharp KA (2005) Heat capacity in proteins. Annu Rev Phys Chem 56:521–548
Oda M, Nakamura H (2000) Thermodynamic and kinetic analyses for understanding sequence-specific DNA recognition. Genes Cells 5:319–326
Tahirov TH, Inoue-Bungo T, Morii H, Fujikawa A, Sasaki M, Kimura K, Shiina M, Sato K, Kumasaka T, Yamamoto M, Ishii S, Ogata K (2001) Structural analyses of DNA recognition by the AML1/Runx-1 Runt domain and its allosteric control by CBFβ. Cell 104:755–767
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera – a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612
Acknowledgments
This study was supported by research grants of Innovative Area, Structural Cell Biology (Number 23121528), and the Strategic Research Foundation Grant-aided Project for Private Universities (Number S1101001) from the Ministry of Education, Sports, Culture, Science and Technology (MEXT) of Japan.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Amano, R., Furukawa, T., Sakamoto, T. (2019). ITC Measurement for High-Affinity Aptamers Binding to Their Target Proteins. In: Ennifar, E. (eds) Microcalorimetry of Biological Molecules. Methods in Molecular Biology, vol 1964. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-9179-2_9
Download citation
DOI: https://doi.org/10.1007/978-1-4939-9179-2_9
Published:
Publisher Name: Humana Press, New York, NY
Print ISBN: 978-1-4939-9178-5
Online ISBN: 978-1-4939-9179-2
eBook Packages: Springer Protocols