Abstract
The distinction between orthologs and paralogs, genes that started diverging by speciation versus duplication, is relevant in a wide range of contexts, most notably phylogenetic tree inference and protein function annotation. In this chapter, we provide an overview of the methods used to infer orthology and paralogy. We survey both graph-based approaches (and their various grouping strategies) and tree-based approaches, which solve the more general problem of gene/species tree reconciliation. We discuss conceptual differences among the various orthology inference methods and databases and examine the difficult issue of verifying and benchmarking orthology predictions. Finally, we review typical applications of orthologous genes, groups, and reconciled trees and conclude with thoughts on future methodological developments.
Adrian M. Altenhoff and Natasha M. Glover are the Joint first authors
You have full access to this open access chapter, Download protocol PDF
Similar content being viewed by others
Key words
1 Introduction
The study of genetic material almost always starts with identifying, within or across species, homologous regions—regions of common ancestry. As we have seen in previous chapters, this can be done at the level of genome segments [1], genes [2], or even down to single residues, in sequence alignments [3]. Here, we focus on genes as evolutionary and functional units. The central premise of this chapter is that it is useful to distinguish between two classes of homologous genes: orthologs, which are pairs of genes that started diverging via evolutionary speciation, and paralogs, which are pairs of genes that started diverging via gene duplication [4] (Fig. 1, Box 1). Originally, the terms and their definition were proposed by Walter M. Fitch in the context of species phylogeny inference, i.e., the reconstruction of the tree of life. He stated “Phylogenies require orthologous, not paralogous, genes” [4]. Indeed, since orthologs arise by speciation, any set of genes in which every pair is orthologous has by definition the same evolutionary history as the underlying species. These days, however, the most frequent motivation for the orthology/paralogy distinction is to study and predict gene function: it is generally believed that orthologs—because they were the same gene in the last common ancestor of the species involved—are likely to have similar biological function. By contrast, paralogs—because they result from duplicated genes that have been retained, at least partly, over the course of evolution—are believed to often differ in function. Consequently, orthologs are of interest to infer function computationally, while paralogs are commonly used to study function innovation.
(a) Simple evolutionary scenario of a gene family with two speciation events (S 1 and S 2) and one duplication event (star). The type of events completely and unambiguously define all pairs of orthologs and paralogs: The frog gene is orthologous to all other genes (they coalesce at S 1). The red and blue genes are orthologs between themselves (they coalesce at S 2), but paralogs between each other (they coalesce at star). (b) The corresponding orthology graph. The genes are represented here by vertices and orthology relationships by edges. The frog gene forms one-to-many orthology with both the human and dog genes, because it is orthologous to more than one sequence in each of these organisms. In such cases, the bi-directional best-hit approach only recovers one of the relations (the highest scoring one). Note that in contrary to BBH, the nonsymmetric BeTs approach—simply taking the best genome-wide hit for each gene regardless of reciprocity—would in the situation of a lost blue human gene infer an incorrect orthologous relation between the blue dog and red human gene
Box 1: Terminology
Homology is a relation between a pair of genes that share a common ancestor. All pairs of genes in the below figure are homologous to each other.

Orthology is a relation defined over a pair of homologous genes, where the two genes have emerged through a speciation event [4]. Example pairs of orthologs are (x 1, y 1) or (x 2, z 1). Orthologs can be further subclassified into one-to-one, one-to-many, many-to-one, and many-to-many orthologs. The qualifiers one and many indicate for each of the two involved genes whether they underwent an additional duplication after the speciation between the two genomes. Hence, the gene pair (x 1, y 1) is an example of a one-to-one orthologous pair, whereas (x 2, z 1) is a many-to-one ortholog relation.
Paralogy is a relation defined over a pair of homologous genes that have emerged through a gene duplication, e.g., (x 1, x 2) or (x 1, y 2).
In-Paralogy is a relation defined over a triplet. It involves a pair of genes and a speciation event of reference. A gene pair is an in-paralog if they are paralogs and duplicated after the speciation event of reference [5]. The pair (x 1, y 2) are in-paralogs with respect to the speciation event S 1.
Out-Paralogy is also a relation defined over a pair of genes and a speciation event of reference. This pair is out-paralogs if the duplication event through which they are related to each other predates the speciation event of reference. Hence, the pair (x 1, y 2) are out-paralogs with respect to the speciation event S 2.
Co-orthology is a relation defined over three genes, where two of them are in-paralogs with respect to the speciation event associated to the third gene. The two in-paralogous genes are said to be co-orthologous to the third (out-group) gene. Thus, x 1 and y 2 are co-orthologs with respect to z 1.
Homoeology is a specific type of homologous relation in a polyploid species, which thus contain multiple “sub-genomes.” This relation describes pairs of genes that originated by speciation and were brought back together in the same genome by allopolyploidization (hybridization) [6]. Thus, in the absence of rearrangement, homoeologs can be thought of as orthologs between sub-genomes.
In this chapter, we first review the main methods used to infer orthology and paralogy, including recent techniques for scaling up algorithms to big data. We then discuss the problem of benchmarking orthology inference. In the last main section, we focus on various applications of orthology and paralogy.
2 Inferring Orthology
Most orthology inference methods can be classified into two major types: graph-based methods and tree-based methods [7]. Methods of the first type rely on graphs with genes (or proteins) as nodes and evolutionary relationships as edges. They infer whether these edges represent orthology or paralogy and build clusters of genes on the basis of the graph. Methods of the second type are based on gene/species tree reconciliation, which is the process of annotating all splits of a given gene tree as duplication or speciation, given the phylogeny of the relevant species. From the reconciled tree, it is trivial to derive all pairs of orthologous and paralogous genes. All pairs of genes which coalesce in a speciation node are orthologs and paralogs if they split at a duplication node. In this section, we present the concepts and methods associated with the two types and discuss the advantages, limitations, and challenges associated with them.
2.1 Graph-Based Methods
Graph-based approaches were originally motivated by the availability of complete genome sequences and the need for efficient methods to detect orthology. They typically run in two phases: a graph construction phase, in which pairs of orthologous genes are inferred (implicitly or explicitly) and connected by edges, and a clustering phase, in which groups of orthologous genes are constructed based on the structure of the graph.
2.1.1 Graph Construction Phase: Orthology Inference
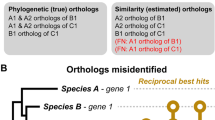
In its most basic form, the graph construction phase identifies orthologous genes by considering pairs of genomes at a time. The main idea is that between any given two genomes, the orthologs tend to be the homologs that diverged least. Why? Because assuming that speciation and duplication are the only types of branching events, the orthologs branched by definition at the latest possible time point—the speciation between the two genomes in question. Therefore, using sequence similarity score as surrogate measure of closeness, the basic approach identifies the corresponding ortholog of each gene through its genome-wide best hit (BeT)—the highest scoring match in the other genome [8]. To make the inference symmetric (as orthology is a symmetric relation), it is usually required that BeTs be reciprocal, i.e., that orthology be inferred for a pair of genes g 1 and g 2 if and only if g 2 is the BeT of g 1 and g 1 is the BeT of g 2 [9]. This symmetric variant, referred to as bi-directional best hit (BBH), has also the merit of being more robust against a possible gene loss in one of the two lineages (Fig. 1).
Inferring orthology from BBH is computationally efficient, because each genome pair can be processed independently and high-scoring alignments can be computed efficiently using dynamic programming [10] or heuristics such as BLAST [11]. Overall, the time complexity scales quadratically in terms of the total number of genes (Box 2). Furthermore, the implementation of this kind of algorithm is simple.
Box 2: Computational Considerations for Scaling to Many Genomes
Time complexity—the amount of time for an algorithm to run as a function of the input—is an important consideration when dealing with big data. This is relevant for inferring orthologs and paralogs due to the massive amounts of sequence data. Thus, it is necessary to consider the time complexity of the inference algorithms, especially when scaling for large and multiple genomes. In computer science, this is commonly denoted in terms of “Big O” notation, which expresses the scaling behavior of the algorithm, up to a constant factor. Below are listed the common time complexities for aspects of some orthology inference algorithms, in order of most efficient to least efficient.
-
Linear time
-
Quadratic time
-
O(n 2): The all-against-all stage central to many orthology algorithms scales quadratically, where n is total number of genes.
-
-
Cubic time
-
O(n 3): The COG database’s graph-based clustering merge triplets of homologs which share a common face until no more can be added.
-
-
NP-complete
-
“Nondeterministic polynomial time,” a large class of algorithms for which no solution in polynomial time is known, (e.g. scaling exponentially with respect to the input size), and thus are impractical. NP-complete problems are typically solved approximately, using heuristics. For instance, maximum likelihood gene tree estimation is NP-complete [14].
-
However, orthology inference by BBH has several limitations, which motivated the development of various improvements (Table 1).
2.1.1.1 Allowing for More Than One Ortholog
Some genes can have more than one orthologous counterpart in a given genome. This happens whenever a gene undergoes duplication after the speciation of the two genomes in question. Since BBH only picks the best hit, it only captures part of the orthologous relations (Fig. 1). The existence of multiple orthologous counterparts is often referred to as one-to-many or many-to-many orthology, depending whether duplication took place in one or both lineages. To designate the copies resulting from such duplications occurring after a speciation of reference, Remm et al. coined the term in-paralogs and introduced a method called InParanoid that improves upon BBH by potentially identifying all pairs of many-to-many orthologs [5]. In brief, their algorithm identifies all paralogs within a species that are evolutionarily closer (more similar) to each other than to the BBH gene in the other genome. This results in two sets of in-paralogs—one for each species—where all pairwise combinations between the two sets are orthologous relations. Alternatively, it is possible to identify many-to-many orthology by relaxing the notion of “best hit” to “group of best hits.” This can be implemented using a score tolerance threshold or a confidence interval around the BBH [23, 34].
2.1.1.2 Evolutionary Distances
Instead of using sequence similarity as a surrogate for evolutionary distance to identify the closest gene(s), Wall et al. proposed to use direct and proper maximum likelihood estimates of the evolutionary distance between pairs of sequences [31]. This estimate of evolutionary distance is based on the number and type of amino acid substitutions between the two sequences. Indeed, previous studies have shown that the highest scoring alignment is often not the nearest phylogenetic neighbor [35]. Building upon this work, Roth et al. showed how statistical uncertainties in the distance estimation can be incorporated into the inference strategy [36].
2.1.1.3 Differential Gene Losses
As discussed above, one of the advantages of BBH over BeT is that by virtue of the bi-directional requirement, the former is more robust to gene losses in one of the two lineages. But if gene losses occurred along both lineages, it can happen that a pair of genes mutually closest to one another is in fact paralogs, simply because both their corresponding orthologs were lost—a situation referred to as “differential gene losses.” Dessimoz et al. [37] presented a way to detect some of these cases by looking for a third species in which the corresponding orthologs have not been lost and thus can act as witnesses of non-orthology.
2.1.2 Clustering Phase: From Pairs to Groups
The graph construction phase yields orthologous relationships between pairs of genes. But this is often not sufficient. Conceptually, information obtained from multiple genes or organisms is often more powerful than that obtained from pairwise comparisons only. In particular, as the use of a third genome as potential witness of non-orthology suggests, a more global view can allow identification and correction of inconsistent/spurious predictions. Practically, it is more intuitive and convenient to work with groups of genes than with a list of gene pairs. Therefore, it is often desirable to cluster orthologous genes into groups.
Tatusov et al. [8] introduced the concept of clusters of orthologous groups (COGs). COGs are computed by using triangles (triplets of genes connected to each other) as seeds and then merging triangles which share a common face, until no more triangle can be added. This clustering can be computed relatively efficient in time O(n 3), where n is the number of genomes analyzed [38]. The stated objective of this clustering procedure is to group genes that have diverged from a single gene in the last common ancestor of the species represented [8]. Practically, they have been found to be useful by many, most notably to categorize prokaryotic genes into broad functional categories.
A different clustering approach was adopted by OrthoMCL, another well-established graph-based orthology inference method [29]. There, groups of orthologs are identified by Markov Clustering [39]. In essence, the method consists in simulating a random walk on the orthology graph, where the edges are weighted according to similarity scores. The Markov Clustering process gives rise to probabilities that two genes belong to the same cluster. The graph is then partitioned according to these probabilities and members of each partition form an orthologous group. These groups contain orthologs and “recent” paralogous genes, where the recency of the paralogs can be somewhat controlled through the parameters of the clustering process.
A third grouping strategy consists in building groups by identifying fully connected subgraphs (called “cliques” in graph theory) [23]. This approach has the merits of straightforward interpretation (groups of genes which are all orthologous to one another) and high confidence in terms of orthology within the resulting groups, due to the high consistency required to form a fully connected subgraph. But it has the drawbacks of being hard to compute (clique finding belongs to the NP-complete class of problems, for which no polynomial time algorithm is known; see Box 2) and being excessively conservative for many applications.
As emerges from these various strategies, there is more than one way orthologous groups can be defined, each with different implications in terms of group properties and applications [40]. In fact, there is an inherent trade-off in partitioning the orthology graph into clusters of genes, because orthology is a non-transitive relation: if genes A and B are orthologs and genes B and C are orthologs, genes A and C are not necessarily orthologs, e.g., consider in Fig. 1 the blue human gene, the frog gene, and the red dog gene. Therefore, if groups are defined as sets of genes in which all pairs of genes are orthologs (as with OMA groups), it is not possible to partition A, B, and C into groups capturing all orthologous relations while leaving out all paralogous relations.
2.1.3 Hierarchical Clustering
More inclusive grouping strategies necessarily lead to orthologs and paralogs within the same group. Nevertheless, it can be possible to control the nature of the paralogs included. For instance, as seen above, OrthoMCL attempts at including only “recent” paralogs in its groups. This idea can be specified more precisely by defining groups with respect to a particular speciation event of interest, e.g., the base of the mammals. Such hierarchical groups are expected to include orthologs and in-paralogs with respect to the reference speciation—in our example all copies that have descended from a single common ancestor gene in the last mammalian common ancestor. Conceptually, hierarchical orthologous groups can be defined as groups of genes that have descended from a single common ancestral gene within a taxonomic range of interest.
Several resources provide hierarchical clustering of orthologous groups. EggNOG [15] and OrthoDB [25], for example, both implement this concept by applying a COG-like clustering method for various taxonomic ranges. Another example, Hieranoid, produces hierarchical groups by using a guide tree to perform pairwise orthology inferences at each node from the leaves to the root—inferring ancestral genomes at each node in the tree [13, 18]. Similarly, OMA GETHOGs is an approach based on an orthology graph of pairwise orthologous gene relations, where hierarchical orthologous groups are formed starting with the most specific taxonomy and incrementally merges them toward the root [21, 22]. Another method, COCO-CL, identifies hierarchical orthologous groups recursively, using correlations of similarity scores among homologous genes [41] and, interestingly, without relying on a species tree. By capturing part of the gene tree structure in the group hierarchies, these methods try in some way to bridge the gap between graph-based and tree-based orthology inference approaches. We now turn our attention to the latter.
2.2 Tree-Based Methods
At their core, tree-based methods infer orthologs on the basis of gene family trees whose internal nodes are labeled as speciation or duplication nodes. Indeed, once all nodes of the gene tree have been inferred as a speciation or duplication event, it is trivial to establish whether a pair of genes is orthologous or paralogous, based on the type of the branching where they coalesce. Such labeling is traditionally obtained by reconciling gene and species trees. In most cases, gene and species trees have different topologies, due to evolutionary events acting specifically on genes such as duplications, losses, lateral transfers, or incomplete lineage sorting [42]. Goodman et al. [43] pioneered research to resolve these incongruences. They showed how the incongruences can be explained in terms of speciation, duplication, and loss events on the gene tree (Fig. 2) and provided an algorithm to infer such events.
Schematic example of the gene/species tree reconciliation. The gene tree and species tree are not compatible. Reconciliation methods resolve the incongruence between the two by inferring speciation, duplication, and losses events on the gene tree. The reconciled tree indicates the most parsimonious history of this gene, constrained to the species tree. The simple representation (bottom right) suggests that the human and frog genes are orthologs and that they are both paralogous to the dog gene
Most tree reconciliation methods rely on a parsimony criterion: the most likely reconciliation is the one which requires the least number of gene duplications and losses. This makes it possible to compute reconciliation efficiently and is tenable as long as duplication and loss events are rare compared to speciation events. In their seminal article, Goodman et al. [43] had already devised their reconciliation algorithm under a parsimony strategy. In the subsequent years, the problem was formalized in terms of a map function between the gene and species trees [44], whose computational cost was conjectured [45], and later proved [12, 46] to coincide with the number of gene duplication and losses. These results yielded highly efficient algorithms, either in terms of asymptotic time complexity [12] or in terms of runtimes on typical problem sizes [47]. With these near-optimal solutions, one might think that the tree reconciliation problem has long been solved. As we shall see in the rest of this section, however, the original formulation of the tree reconciliation problem has several limitations in practice, which have stimulated the development of various refinements to overcome them (Table 2).
2.2.1 Unresolved Species Tree
A first problem ignored by most early reconciliation algorithms lies in the uncertainty often associated with the species tree, which these methods assume as correct and heavily rely upon.
One way of dealing with the uncertainties is to treat unresolved parts of the species tree as multifurcating nodes (also known as soft polytomies). By doing so, the reconciliation algorithm is not forced to choose for a specific type of evolutionary event in ambiguous regions of the tree. This approach is, for instance, implemented in TreeBeST [52] and used in the Ensembl Compara project [53].
Alternatively, Heijden et al. [57] demonstrated that it is often possible to infer speciation and duplication events on a gene tree without knowledge of the species tree. Their approach, which they call species overlap, identifies for a given split the species represented in the two subtrees induced by the split. If at least one species has genes in both subtrees, a duplication event is inferred; else a speciation event is inferred. In fact, this approach is a special case of soft polytomies where all internal nodes have been collapsed. Thus, the only information needed for this approach is a rooted gene tree. Since then, this approach has been adopted in other projects, such as PhylomeDB [59].
2.2.2 Rooting
The classical reconciliation formulation requires both gene and species trees to be rooted. But most models of sequence evolution are time reversible and thus do not allow to infer the rooting of the reconstructed gene tree. One sensible solution is to root a gene tree so that it minimizes the number of duplication events [62]. Thus, this method uses the parsimony principle for both rooting and reconciliation. For cases of multiple optimal rootings, ties can be broken by selecting the tree that minimizes the tree height [63] or by picking the rooting which minimizes the number of gene losses [61].
Another approach is to place the root at the “center of the tree”—also known as “midpoint rooting” [58]. The idea of this method goes back to Farris [64] and is motivated by the concept of a molecular clock. But for most gene families, assuming a constant rate of evolution is inappropriate [65, 66], and thus this approach is not used widely. A newly introduced refinement based on minimizing average deviations among children nodes holds promise of being more robust [67] but still relies on a molecular clock assumption.
For the species tree, the most common and reliable way of rooting trees is by identifying an outgroup species. PhylomeDB uses genes from outgroup species to root gene trees [59]. One main potential problem with this approach is that in many situations, it can be difficult to identify a suitable outgroup. For example, in analysis covering all kingdoms of life, an outgroup species may not be available, or the relevant genes might have been lost [68]. A suitable out-group needs to be close enough to allow for reliable sequence alignment, yet it must have speciated clearly before any other species separated. Furthermore, ancient duplications can cause outgroup species to carry in-group genes. These difficulties make this approach more challenging for automated, large-scale analysis [69].
2.2.3 Gene Tree Uncertainty
Another assumption made in the original tree reconciliation problem is the (topological) correctness of the gene tree. But it has been shown that this assumption is commonly violated, often due to finite sequence lengths, taxon sampling [70, 71], or gene evolution model violations [72]. On the other hand, techniques of expressing uncertainties in gene tree reconstruction via support measures, e.g., bootstrap values, have become well established. Storm and Sonnhammer [58] as well as Zmasek and Eddy [63] independently suggested to extend the bootstrap procedure to reconciliation, thereby reducing the dependency of the reconciliation procedure on any one gene tree while providing a measure of support of the inferred speciation/duplication events. The downsides of using the bootstrap are the high computational costs and interpretation difficulties associated with it [73].
Similarly to how unresolved species tree can be handled, unresolved parts of the gene tree can also be collapsed into multifurcating nodes. For instance, HOGENOM [55] and Softparsmap [61] collapse branches with low bootstrap support values.
A third way of tackling this problem consists in simultaneously solving both the gene tree reconstruction and reconciliation problems [74]. They use the parsimony criterion of minimizing the number of duplication events to improve on the gene tree itself. This is achieved by rearranging the local gene tree topology of regions with low bootstrap support such that the number of duplications and losses is further reduced.
2.2.4 Parsimony vs. Likelihood
All the approaches mentioned so far try to minimize the number of gene duplication events. This is generally justified by a parsimony argument, which assumes that gene duplications and losses are rare events. But what if this assumption is frequently violated? Little is known about duplication and loss rates in general [75], but there is strong evidence for historical periods with high gene duplication occurrence rates [76] or gene families specifically prone to massive duplications (e.g., olfactory receptor, opsins, serine/threonine kinases, etc.)
Motivated by this reasoning, Arvestad et al. introduced the idea of a probabilistic model for tree reconciliation [49]. They used a Bayesian approach to estimate the posterior probabilities of a reconciliation between a given gene and species tree using Markov chain Monte Carlo (MCMC) techniques. Arvestad et al. [49] modeled gene duplication and loss events through a birth-death process [77]. In the subsequent years, they refined their method to also model sequence evolution and substitution rates in a unified framework called gene sequence evolution model with iid rates (GSR) [49, 50].
Perhaps the biggest problem with the probabilistic approach is that it is not clear how well the assumptions of their model (the birth-death process with fixed parameters) relate to the true process of gene duplication and gene loss. Doyon et al. [78] compared the maximum parsimony reconciliation trees from 1278 fungi gene families to the probabilistically reconciled trees using gene birth/death rates fitted from the data. They found that in all but two cases, the maximum parsimony scenario corresponds to the most probable one. This remarkably high level of consistency indicates that in terms of the accuracy of the “best” reconciliation, there is little to gain from using a likelihood approach over the parsimony criterion of minimizing the number of duplication events. But how this result generalizes to other datasets has yet to be investigated.
2.3 Graph-Based vs. Tree-Based: Which Is Better?
Given the two fundamentally different paradigms in orthology inference that we reviewed in this section, one can wonder which is better. Conceptually, tree reconciliation methods have several advantages. In terms of inference, by considering all sequences from all species at the same time, it can also be expected that they can extract more information from the sequences. This in turn should translate into higher statistical power. In terms of their output, reconciled gene trees provide the user more information than pairs or groups of orthologs. For example, the trees display the order of duplication and speciation events, as well as evolutionary distances between these events. In practice, however, these methods have the disadvantage of having much higher computational complexity than their graph-based counterparts. Furthermore, the two approaches are in practice often not that strictly separated. Tree-based methods often start with a graph-based clustering step to identify families of homologous genes. Conversely, several hierarchical grouping algorithms also rely on species trees in their inference.
Thus, it is difficult to make general statements about the relative performance of the two classes of inference methods. One solution that can leverage the unique abilities of both tree-based and graph-based methods is to combine several independent orthology inference methods into one. We discuss this technique in the next section.
3 Meta-methods
In recent years a new class of orthology inference tools has emerged which attempts to make the most out of multiple orthology prediction algorithms—meta-methods. These are approaches which combine several individual and distinct methods in order to produce more robust orthology predictions. These meta-methods are able to take advantage of the standardized formats of output which has been a goal of the orthology community [79], as well as the many new and well-established methods out there.
Generally, meta-methods assign a confidence score to a given predicted orthologous relation. In its most basic form, more weight is given to orthologs predicted by the most methods. Some examples include methods which simply take the intersection of several methods, such as GET_HOMOLOGUES [80], COMPARE [81], HCOP [82], and DIOPT [83]. These methods maintain a high level of precision, but since they are based on intersections, they necessarily have a lower recall.
Additionally, post-processing techniques can be used to build upon the base of orthologs found by several methods—thus assigning more sequences as orthologs and improving performance. For example, MOSAIC (Multiple Orthologous Sequence Analysis and Integration by Cluster optimization) [84] uses an iterative graph-based optimization approach that works on ortholog sets predicted by several independent methods. MOSAIC captures orthologs which are missed by some individual methods, producing a 1.6-fold increase in the number of orthologs detected. Another example is the MARIO software, which looks for the intersection of several different orthology methods as seed groups and then progressively adds unassigned proteins to the groups based on HMM profiles [85]. MetaPhOrs’ approach integrates phylogenetic and homology information derived from different databases [86]. They demonstrate that the number of independent sources from which an orthology prediction is made, as well as the level of consistency across predictions, can be used as confidence scores.
So far the previously mentioned meta-methods combine independent orthology prediction algorithms and give a higher score based on the more algorithms which predict a given orthologous relation. However, another emerging approach is to use machine learning techniques to recognize patterns among several different orthology inference methods. With this, one can predict previously unknown high-confidence orthologs. WORMHOLE is a tool which uses the information from 17 different orthology prediction methods to train support vector machine classifiers for predicting least diverged orthologs [87]. WORMHOLE was able to strongly re-predict least diverged orthologs in the reference set and also predict previously unclassified orthologous genes.
The type of meta-approach and its associated stringency depends on what the user is going after. For example, if the goal is to get very-high-confidence groups, methods which only combine for the intersection without trying to add more orthologs may be preferable. Studies requiring both high precision and recall may be better suited to use the meta-methods which use post-processing or machine learning to predict orthologs. And as with all methods, it is important to understand which clades the method has been benchmarked in and which orthology tools have been combined. For example, if several methods have the same bias, one will just propagate the bias and end up with a false sense of security because the methods are not independent.
4 Scaling to Many Genomes
In terms of orthology inference, the abundance of genomes now available has resulted in an emphasis on driving down computational processing time via efficient algorithms. When inferring orthology for many genomes, the bottleneck is generally the all-against-all computations—aligning the proteins in every genome against the proteins in every other genome. This is the first step of nearly all graph-based methods. The all-against-all computation has an O(n 2) runtime, meaning it scales quadratically with the number of genomes analyzed (Box 2).
So far, two main techniques for scaling orthology prediction to many genomes have emerged. The first approach is by making the all-against-all comparisons faster. Because comparisons are independent of each other, the most obvious way of doing this is by taking advantage of a high-performance computing cluster, as this is an embarrassingly parallel computing problem. Many methods have implemented this, such as Hieranoid [13], PorthoMCL [88], or OMA [22]. Another way to save time on the all-against-all comparisons is by using very fast algorithms for the homology search. For example, preliminary results of SonicParanoid showed 160–750× speedup of orthology inference compared to InParanoid [89]. Innovations in alignment algorithms with methods such as DIAMOND [90] or MMSeq2 [91] have the potential to greatly reduce the time to do the all-against-all comparisons.
A second approach to efficiently scale up orthology inference to many genomes is by simply avoiding doing the entire all-against-all comparisons. This makes sense, since a significant amount of time is spent comparing unrelated gene pairs. For example, it is possible to avoid aligning many unrelated pairs by exploiting the transitive property of homology. Wittwer et al. [92] did this by first building clusters of homologous sequences with one representative sequence per cluster and subsequently performing the all-against-all within each cluster. Hieranoid avoids unnecessary all-against-all comparisons by using a species tree as a guide, reducing the number of comparisons to N − 1 for N genomes, scaling linearly rather than quadratically [18]. Another way to avoid all-by-all comparison is by using a mapping strategy, whereby new proteomes are mapped onto precomputed orthologous groups. This strategy has been successfully implemented with the eggNOG database—each sequence in a new proteome is mapped to a precomputed orthologous cluster based on hidden Markov models. Then, orthology relations and function are transferred to the new sequence from the best matching sequence in the database [93].
5 Benchmarking Orthology
Assessing the quality of orthology predictions is important but difficult. The main challenge is that the precise evolutionary history of entire genomes is largely unknown and thus, predictions can only be validated indirectly, using surrogate measures. To be informative, such measures need to strongly correlate with orthology/paralogy. At the same time, they should be independent from the methods used in the orthology inference process. Concretely, this means that the orthology inference is not based on the surrogate measure and the surrogate measure is not derived from orthology/paralogy.
5.1 Benchmarking Approaches
Several ways of benchmarking orthology inference have been developed in the past years. In the next sections, we go over the main approaches, bringing attention to the advantages and limitations to each.
5.1.1 Functional Conservation
The first surrogate measures proposed revolved around conservation of function [94]. This was motivated by the common belief that orthologs tend to have conserved function, while paralogs tend to have different functions. Indeed, orthologs tend to be more conserved than paralogs in terms of GO annotation similarity [95]. Thus, “for a given evolutionary distance, more accurate orthology inference is likely to be correlated with more functionally similar gene pairs.” Hulsen et al. [94] assessed the quality of ortholog predictions in terms of conservation of co-expression levels, domain annotation, and protein-protein interaction partners. Additionally, Altenhoff et al. [96] used similarity of experimentally validated GO annotations as well as Enzyme Commission (EC) numbers as a functional benchmark. Functional benchmarks have an advantage in that many researchers are interested in orthology because they want to find functionally conserved genes, thus making functional tests important for assessing different inference methods. The main limitation of these measures is that it is not so clear how much they correlate with orthology/paralogy. Indeed, it has been argued that the difference in function conservation trends between orthologs and paralogs might be much smaller than commonly assumed and indeed many examples are known of orthologs that have dramatically different functions [97].
5.1.2 Gene Neighborhood Conservation
The fraction of orthologs that have neighboring genes being orthologs themselves is an indicator of consistency and therefore to some extent also of quality of orthology predictions [94]. Although synteny has been used as part of the orthology inference for several algorithms, to date it has not been used as part of large-scale benchmarking efforts. One possible problem is that gene neighborhood can be conserved among paralogs, such as those resulting from whole-genome duplications. Furthermore, some methods use gene neighborhood conservation to help in their inference process, which can bias the assessment done on such measures (principle of independence stated above).
5.1.3 Species Tree Discordance Test
The quality of ortholog predictions can also be assessed based on phylogeny. By definition, the tree relating a set of genes all orthologous to one another only contains speciation splits and has the same topology as the underlying species. We introduced a benchmarking protocol that quantifies how well the predictions from various orthology inference methods agree with undisputed species tree topologies [96, 98]. Thus, the species tree discordance test judges the accuracy of ortholog predictions based on the correctness of the species tree which can be constructed from them. The advantage of this measure is that by virtue of directly ensuing from the definition of orthology, it correlates strongly with it and thus satisfies the first principle. However, the second principle, independence from the inference process, is not satisfied with methods relying on the species tree—typically all reconciliation methods but also most graph-based methods producing hierarchical groups. In such cases, interpretation of the results must be done carefully.
5.1.4 Gold Standard Gene Tree Test
High-quality reference gene trees can also be used to assess orthology inferences. For this, one compares the pairs of orthologs from a given method to pairs of orthologs derived from these expertly curated gene trees [40, 99]. One drawback of this benchmark is that it is limited by the ability to curate the phylogeny—if the evolutionary history of the gene family is ambiguous, the resulting reference tree will unavoidably have mistakes. Another limitation is the small size of most benchmarks of this type. This casts doubts on their generalizability and makes them prone to overfitting.
5.1.5 Subtree Consistency Test
For inference methods based on reconciliation between gene and species trees, Vilella et al. [53] proposed a different phylogeny-based assessment scheme. For any duplication node of the labeled gene tree, a consistency score is computed, which captures the balance of the species found in the two subtrees. Unbalanced nodes correspond to an evolutionary scenario involving extensive gene losses and therefore, under the principle of parsimony, are less likely to be correct. Given that studies to date tend to support the adequacy of the parsimony criterion in the context of gene family dynamics (Subheading 2.2.4), it can be expected that this metric correlates highly with correct orthology/paralogy assignments. However, since virtually all tree-based methods themselves incorporate this very criterion in their objective function (i.e., minimizing the number of gene duplications and losses), the principle of independence is violated, and thus the adequacy of this measure is questionable.
5.1.6 Latent Class Analysis
Chen et al. [100] proposed a purely statistical benchmark based on latent class analysis (LCA). Given the absence of a definitive answer on whether two given genes are orthologs, the authors argue that by looking at the agreement and disagreement of predictions made by several inference methods on a common dataset, one can estimate the reliability of individual predictors. More precisely, LCA is a statistical technique that computes maximum likelihood estimates of sensitivity and specificity rates for each orthology inference methods, given their predictions and given an error model. This is attractive, because it does not depend on any surrogate measure. However, the results depend on the error model assumed. Thus, we are of the opinion that LCA merely shifts the problem of assessing orthology to the problem of assessing an error model of various orthology inference methods.
5.1.7 Simulated Genomes
Finally, simulated data can be used in benchmarking. By this, the precise evolutionary history of a genome can be validated, in terms of gene duplication, insertion, deletion, and lateral gene transfer [101]. Knowing for certain all aspects of the simulated genomes gives an advantage over assessments based on empirical data, where the true evolutionary history is unknown. On the other hand, how well the simulated data reflect “real” data is debatable.
5.2 Orthology Benchmarking Service
The orthology benchmarking service is a web-based platform for which users can upload their ortholog predictions and run them through a variety of benchmarks. The user must use quest for orthologs (QFO) reference proteome set, which is a set of 66 genomes that covers a diverse set of species across all domains [79], to infer pairwise or groups of orthologs. Several phylogenetic and function-based benchmarks are automatically run on the uploaded data, and then summary statistics of the results of each benchmark are reported. The user can compare their method’s performance with that of other well-known orthology inference algorithms and choose to make theirs public as well. For each benchmark, a precision-recall curve is reported, allowing for ease of comparison and evaluation of individual inference techniques. Because of the range of benchmarking tests and publicly available methods for comparison, the benchmarking service is useful for both users, who can check which methods work well for their particular problem and for method developers. The orthology benchmarking service can be accessed at http://orthology.benchmarkservice.org.
5.3 Conclusions on Benchmarking
Overall, it becomes apparent that there is no “magic bullet” strategy for orthology benchmarking, as each approach discussed here has its limitations (though some limitations are more serious than others). Nevertheless, comparative studies based on these various benchmarking measures have reported surprisingly consistent findings [40, 94, 96, 98, 100]: these assessments generally observe that there is a trade-off between accuracy and coverage and most common databases are situated on a Pareto frontier. The various assessments concur that the “best” orthology approach is highly dependent on the various possible applications of orthology.
6 Applications
As we have seen so far, there is a large diversity in the methods for orthology inference. The main reason is that, although the methods discussed here all infer orthology as part of their process, many of them have been developed for different reasons and have different ultimate goals. Unfortunately, this is often not mentioned explicitly and tends to be a source of confusion. In this section, we review some of these ultimate goals and discuss which methods and representation of orthology are better suited to address them and why.
As mentioned in the introduction, most interest for orthology is in the context of function prediction and is largely based on the belief that orthologs tend to have conserved function. A conservative approach consists in propagating function between one-to-one orthologs, i.e., pairs of orthologous genes that have not undergone gene duplication since they diverged from one another. Several orthology databases directly provide one-to-one orthology predictions. But even with those that do not, it might still be possible to obtain such predictions, for instance, by selecting hierarchical groups containing at most one sequence in each species or by extracting from reconciled trees’ subtrees with no duplication. A more sophisticated approach consists in propagating gene function annotations across genomes on the basis of the full reconciled gene tree. Thomas et al. [102], for instance, proposed a way to assign gene function to uncharacterized proteins using a gene tree and a hidden Markov model (HMM) among gene families. Engelhardt et al.. [103] developed a Bayesian model of function change along reconciled gene trees and showed that their approach significantly improves upon several methods based on pairwise gene function propagation. Ensembl Compara [53] and Panther [102] are two major databases providing reconciled gene trees.
Since Darwin, one traditional question in biology has always been how species are related to each other. As we recall in the introduction of this chapter, Fitch’s original motivation for defining orthology was phylogenetic inference. Indeed, the gene tree reconstructed from a set of genes which are all orthologous to each other should by definition be congruent to the species tree. OMA Groups (OMA) have this characteristic and, crucially, are constructed without help of a species tree.
Yet another application associated with orthology are general alignments between genomes, e.g., protein-protein interaction (PPI) network alignments or whole-genome alignments. Finding an optimal PPI network alignment between two genomes on the basis of the network topology alone is a computationally hard problem (i.e., it is an instance of the subgraph isomorphism problem which is NP-complete [104]). Orthology is often used as heuristic to constrain the mapping of the corresponding genes between the two networks and thus to reduce the problem complexity of aligning networks [105]. For whole-genome alignments, people most often use homologous regions and use orthologs as anchor points [106]. These types of application typically rely on ortholog predictions between pairs of genomes, as provided, e.g., by InParanoid [5] or OMA [23].
7 Conclusions and Outlook
The distinction between orthologs and paralogs is at the heart of many comparative genomic studies and applications. The original and generally accepted definition of orthology is based on the evolutionary history of pairs of genes. By contrast, there is a considerable diversity in how groups of orthologs are defined. These differences largely stem from the fact that orthology is a non-transitive relation and therefore, dividing genes into orthologous groups will either miss or wrongly include orthologous relations. This makes it important and worthwhile to identify the type of orthologous group best suited for a given application.
Regarding inference methods, while most approaches can be ordered into two fundamental paradigms—graph-based and tree-based—the difference between the two is shrinking, with graph-based methods increasingly striving to capture more of the evolutionary history. On the other hand, the rapid pace at which new genomes are sequenced limits the applicability of tree-based methods, computationally more demanding.
Benchmarking this large variety of methods remains a hard problem—from a conceptual point as described above but also because of very practical challenges such as heterogeneous data formats, genome versions, or gene identifiers. This has been recognized by the research community and has led to the development of the QFO consortium benchmarking service [96].
Looking forward, we see potential in extending the current model of gene evolution, which is limited to speciation, duplication, and loss events. Indeed, nature is often much more complicated. For instance, lateral gene transfer (LGT) is believed to be a major mode of evolution in prokaryotes. While there has been several attempts at extending tree reconciliation algorithms to detecting LGT [107, 108], the problem is largely unaddressed in typical orthology resources [109]. Another relevant evolutionary process omitted by most methods is whole-genome duplications (WGD). Even though WGD events act jointly on all gene families, with few exceptions [110, 111], most methods consider each gene family independently.
Overall, the orthology/paralogy dichotomy has proved to be useful but also inherently limited. Reducing the whole evolutionary history of homologous genes into binary pairwise relations is bound to be a simplification—and at times an oversimplification. The shift toward hierarchical orthologous groups is thus a promising step toward capturing more features of the evolutionary history of genes. Yet further development will still be needed, as we are nowhere close to grasp the formidable complexity of gene evolution across the full diversity of life.
8 Exercises
Assume the following evolutionary scenario

where duplications are depicted as star and all other splits are speciations.
-
Problem #1: Draw the corresponding orthology graph, where the vertices correspond to the observed genes and the edges indicate orthologous relations between them.
-
Problem #2: Apply the following two clustering methods on your orthology graph. First, reconstruct all the maximal fully connected subgraphs (cliques) that can be found. Second, reconstruct the COGs. COGs are built by merging triangles of orthologs whenever they share a common face. Remember that in both methods, a gene can only belong to a one cluster.
References
Dewey CN (2012) Whole-genome alignment. Methods Mol Biol 855:237–257
Alioto T (2012) Gene prediction. In: Anisimova M (ed) Evolutionary genomics: statistical and computational methods, vol 1. Humana, Totowa, NJ, pp 175–201
Löytynoja A (2012) Alignment methods: strategies, challenges, benchmarking, and comparative overview. In: Anisimova M (ed) Evolutionary genomics: statistical and computational methods, vol 1. Humana, Totowa, NJ, pp 203–235
Fitch WM (1970) Distinguishing homologous from analogous proteins. Syst Zool 19:99–113
Remm M, Storm CEV, Sonnhammer ELL (2001) Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J Mol Biol 314:1041–1052
Glover NM, Redestig H, Dessimoz C (2016) Homoeologs: what are they and how do we infer them? Trends Plant Sci 21:609–621
Kuzniar A, van Ham RCHJ, Pongor S et al (2008) The quest for orthologs: finding the corresponding gene across genomes. Trends Genet 24:539–551
Tatusov RL, Koonin EV, Lipman DJ (1997) A genomic perspective on protein families. Science 278:631–637
Overbeek R, Fonstein M, D’Souza M et al (1999) The use of gene clusters to infer functional coupling. Proc Natl Acad Sci U S A 96:2896–2901
Smith TF, Waterman MS (1981) Identification of common molecular subsequences. J Mol Biol 147:195–197
Altschul SF, Madden TL, Schäffer AA et al (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Zhang L (1997) On a Mirkin-Muchnik-Smith conjecture for comparing molecular phylogenies. J Comput Biol 4:177–187
Schreiber F, Sonnhammer ELL (2013) Hieranoid: hierarchical orthology inference. J Mol Biol 425:2072–2081
Chor B, Tuller T (2005) Maximum likelihood of evolutionary trees is hard. In: Proceedings of the 9th annual international conference on research in computational molecular biology. Springer, Berlin, pp 296–310
Jensen LJ, Julien P, Kuhn M et al (2008) eggNOG: automated construction and annotation of orthologous groups of genes. Nucleic Acids Res 36:D250–D254
Muller J, Szklarczyk D, Julien P et al (2010) eggNOG v2.0: extending the evolutionary genealogy of genes with enhanced non-supervised orthologous groups, species and functional annotations. Nucleic Acids Res 38:D190–D195
Huerta-Cepas J, Szklarczyk D, Forslund K et al (2016) eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res 44:D286–D293
Kaduk M, Sonnhammer E (2017) Improved orthology inference with Hieranoid 2. Bioinformatics 33:1154–1159
Ostlund G, Schmitt T, Forslund K et al (2010) InParanoid 7: new algorithms and tools for eukaryotic orthology analysis. Nucleic Acids Res 38:D196–D203
Sonnhammer ELL, Östlund G (2015) InParanoid 8: orthology analysis between 273 proteomes, mostly eukaryotic. Nucleic Acids Res 43:D234–D239
Altenhoff AM, Gil M, Gonnet GH et al (2013) Inferring hierarchical orthologous groups from orthologous gene pairs. PLoS One 8:e53786
Train C-M, Glover NM, Gonnet GH et al (2017) Orthologous Matrix (OMA) algorithm 2.0: more robust to asymmetric evolutionary rates and more scalable hierarchical orthologous group inference. Bioinformatics 33:i75–i82
Dessimoz C, Cannarozzi G, Gil M et al (2005) OMA, a comprehensive, automated project for the identification of orthologs from complete genome data: introduction and first achievements. In: Comparative genomics. Springer, Berlin, pp 61–72
Altenhoff AM, Schneider A, Gonnet GH et al (2011) OMA 2011: orthology inference among 1000 complete genomes. Nucleic Acids Res 39:D289–D294
Kriventseva EV, Rahman N, Espinosa O et al (2008) OrthoDB: the hierarchical catalog of eukaryotic orthologs. Nucleic Acids Res 36:D271–D275
Zdobnov EM, Tegenfeldt F, Kuznetsov D et al (2017) OrthoDB v9.1: cataloging evolutionary and functional annotations for animal, fungal, plant, archaeal, bacterial and viral orthologs. Nucleic Acids Res 45:D744–D749
Linard B, Thompson JD, Poch O et al (2011) OrthoInspector: comprehensive orthology analysis and visual exploration. BMC Bioinform 12:11
Linard B, Allot A, Schneider R et al (2015) OrthoInspector 2.0: software and database updates. Bioinformatics 31:447–448
Li L, Stoeckert CJ Jr, Roos DS (2003) OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13:2178–2189
Chen F, Mackey AJ, Stoeckert CJ Jr et al (2006) OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res 34:D363–D368
Wall DP, Fraser HB, Hirsh AE (2003) Detecting putative orthologs. Bioinformatics 19:1710–1711
DeLuca TF, Wu I-H, Pu J et al (2006) Roundup: a multi-genome repository of orthologs and evolutionary distances. Bioinformatics 22:2044–2046
DeLuca TF, Cui J, Jung J-Y et al (2012) Roundup 2.0: enabling comparative genomics for over 1800 genomes. Bioinformatics 28:715–716
Fulton DL, Li YY, Laird MR et al (2006) Improving the specificity of high-throughput ortholog prediction. BMC Bioinform 7:270
Koski LB, Golding GB (2001) The closest BLAST hit is often not the nearest neighbor. J Mol Evol 52:540–542
Roth ACJ, Gonnet GH, Dessimoz C (2008) Algorithm of OMA for large-scale orthology inference. BMC Bioinform 9:518
Dessimoz C, Boeckmann B, Roth ACJ et al (2006) Detecting non-orthology in the COGs database and other approaches grouping orthologs using genome-specific best hits. Nucleic Acids Res 34:3309–3316
Kristensen DM, Kannan L, Coleman MK et al (2010) A low-polynomial algorithm for assembling clusters of orthologous groups from intergenomic symmetric best matches. Bioinformatics 26:1481–1487
Van Dongen SM (2001) Graph clustering by flow simulation. PhD thesis, University of Utrecht
Boeckmann B, Robinson-Rechavi M, Xenarios I et al (2011) Conceptual framework and pilot study to benchmark phylogenomic databases based on reference gene trees. Brief Bioinform 12:423–435
Jothi R, Zotenko E, Tasneem A et al (2006) COCO-CL: hierarchical clustering of homology relations based on evolutionary correlations. Bioinformatics 22:779–788
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Goodman M, Czelusniak J, Moore GW et al (1979) Fitting the gene lineage into its species lineage, a Parsimony strategy illustrated by cladograms constructed from globin sequences. Syst Zool 28:132–163
Page RDM (1994) Maps between trees and cladistic analysis of historical associations among genes, organisms, and areas. Syst Biol 43:58–77
Mirkin B, Muchnik I, Smith TF (1995) A biologically consistent model for comparing molecular phylogenies. J Comput Biol 2:493–507
Eulenstein O (1997) A linear time algorithm for tree mapping. Arbeitspapiere der GMD No. 1046, St
Zmasek CM, Eddy SR (2001) A simple algorithm to infer gene duplication and speciation events on a gene tree. Bioinformatics 17:821–828
Poptsova MS, Gogarten JP (2007) BranchClust: a phylogenetic algorithm for selecting gene families. BMC Bioinform 8:120
Arvestad L, Berglund A-C, Lagergren J et al (2003) Bayesian gene/species tree reconciliation and orthology analysis using MCMC. Bioinformatics 19(Suppl 1):i7–i15
Åkerborg Ö, Sennblad B, Arvestad L et al (2009) Simultaneous Bayesian gene tree reconstruction and reconciliation analysis. Proc Natl Acad Sci U S A 106:5714–5719
Ullah I, Sjöstrand J, Andersson P et al (2015) Integrating sequence evolution into probabilistic orthology analysis. Syst Biol 64:969–982
Li H, Coghlan A, Ruan J et al (2006) TreeFam: a curated database of phylogenetic trees of animal gene families. Nucleic Acids Res 34:D572–D580
Vilella AJ, Severin J, Ureta-Vidal A et al (2009) EnsemblCompara GeneTrees: complete, duplication-aware phylogenetic trees in vertebrates. Genome Res 19:327–335
Herrero J, Muffato M, Beal K et al (2016) Ensembl comparative genomics resources. Database 2016:bav096
Dufayard J-F, Duret L, Penel S et al (2005) Tree pattern matching in phylogenetic trees: automatic search for orthologs or paralogs in homologous gene sequence databases. Bioinformatics 21:2596–2603
Penel S, Arigon A-M, Dufayard J-F et al (2009) Databases of homologous gene families for comparative genomics. BMC Bioinform 10(Suppl 6):S3
van der Heijden RTJM, Snel B, van Noort V et al (2007) Orthology prediction at scalable resolution by phylogenetic tree analysis. BMC Bioinform 8:83
Storm CEV, Sonnhammer ELL (2002) Automated ortholog inference from phylogenetic trees and calculation of orthology reliability. Bioinformatics 18:92–99
Huerta-Cepas J, Dopazo H, Dopazo J et al (2007) The human phylome. Genome Biol 8:R109
Huerta-Cepas J, Capella-Gutiérrez S, Pryszcz LP et al (2014) PhylomeDB v4: zooming into the plurality of evolutionary histories of a genome. Nucleic Acids Res 42:D897–D902
Berglund-Sonnhammer A-C, Steffansson P, Betts MJ et al (2006) Optimal gene trees from sequences and species trees using a soft interpretation of parsimony. J Mol Evol 63:240–250
Hallett MT, Lagergren J (2000) New algorithms for the duplication-loss model. In: Proceedings of the fourth annual international conference on computational molecular biology. ACM, New York, NY, pp 138–146
Zmasek CM, Eddy SR (2002) RIO: analyzing proteomes by automated phylogenomics using resampled inference of orthologs. BMC Bioinform 3:14
Farris JS (1972) Estimating phylogenetic trees from distance matrices. Am Nat 106:645–668
Avise JC, Bowen BW, Lamb T et al (1992) Mitochondrial DNA evolution at a turtle’s pace: evidence for low genetic variability and reduced microevolutionary rate in the Testudines. Mol Biol Evol 9:457–473
Ayala FJ (1999) Molecular clock mirages. Bioessays 21:71–75
Tria FDK, Landan G, Dagan T (2017) Phylogenetic rooting using minimal ancestor deviation. Nat Ecol Evol 1:193
Huelsenbeck JP, Bollback JP, Levine AM (2002) Inferring the root of a phylogenetic tree. Syst Biol 51:32–43
Tarrıo R, Rodrıguez-Trelles F, Ayala FJ (2000) Tree rooting with outgroups when they differ in their nucleotide composition from the ingroup: the Drosophila saltans and Willistoni groups, a case study. Mol Phylogenet Evol 16:344–349
Graybeal A (1998) Is it better to add taxa or characters to a difficult phylogenetic problem? Syst Biol 47(1):9–17
Rokas A, Williams BL, King N et al (2003) Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature 425:798–804
Yang Z, Goldman N, Friday A (1994) Comparison of models for nucleotide substitution used in maximum-likelihood phylogenetic estimation. Mol Biol Evol 11:316–324
Anisimova M, Gascuel O (2006) Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst Biol 55:539–552
Durand D, Halldórsson BV, Vernot B (2006) A hybrid micro-macroevolutionary approach to gene tree reconstruction. J Comput Biol 13:320–335
Lynch M, Conery JS (2000) The evolutionary fate and consequences of duplicate genes. Science 290:1151–1155
Robinson-Rechavi M, Marchand O, Escriva H et al (2001) Euteleost fish genomes are characterized by expansion of gene families. Genome Res 11:781–788
Kendall DG (1948) On the generalized “birth-and-death” process. Ann Math Stat 19:1–15
Doyon J-P, Hamel S, Chauve C (2012) An efficient method for exploring the space of gene tree/species tree reconciliations in a probabilistic framework. IEEE/ACM Trans Comput Biol Bioinform 9:26–39
Gabaldón T, Dessimoz C, Huxley-Jones J et al (2009) Joining forces in the quest for orthologs. Genome Biol 10:403
Contreras-Moreira B, Vinuesa P (2013) GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Appl Environ Microbiol 79:7696–7701
Salgado D, Gimenez G, Coulier F et al (2008) COMPARE, a multi-organism system for cross-species data comparison and transfer of information. Bioinformatics 24:447–449
Eyre TA, Wright MW, Lush MJ et al (2007) HCOP: a searchable database of human orthology predictions. Brief Bioinform 8:2–5
Hu Y, Flockhart I, Vinayagam A et al (2011) An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinform 12:357
Maher MC, Hernandez RD (2015) Rock, paper, scissors: harnessing complementarity in ortholog detection methods improves comparative genomic inference. G3 5:629–638
Pereira C, Denise A, Lespinet O (2014) A meta-approach for improving the prediction and the functional annotation of ortholog groups. BMC Genomics 15(Suppl 6):S16
Pryszcz LP, Huerta-Cepas J, Gabaldón T (2011) MetaPhOrs: orthology and paralogy predictions from multiple phylogenetic evidence using a consistency-based confidence score. Nucleic Acids Res 39:e32
Sutphin GL, Mahoney JM, Sheppard K et al (2016) WORMHOLE: novel least diverged ortholog prediction through machine learning. PLoS Comput Biol 12:e1005182
Tabari E, Su Z (2017) PorthoMCL: parallel orthology prediction using MCL for the realm of massive genome availability. Big Data Anal 2:4
Cosentino S, Iwasaki W (2018) SonicParanoid: extremely fast, accurate, and easy orthology inference. Bioinformatics. https://doi.org/10.1093/bioinformatics/bty631
Buchfink B, Xie C, Huson DH (2015) Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60
Steinegger M, Söding J (2017) MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat Biotechnol 35(11):1026–1028
Wittwer LD, Piližota I, Altenhoff AM et al (2014) Speeding up all-against-all protein comparisons while maintaining sensitivity by considering subsequence-level homology. PeerJ 2:e607
Huerta-Cepas J, Forslund K, Coelho LP et al (2017) Fast genome-wide functional annotation through orthology assignment by eggNOG-Mapper. Mol Biol Evol 34:2115–2122
Hulsen T, Huynen MA, de Vlieg J et al (2006) Benchmarking ortholog identification methods using functional genomics data. Genome Biol 7:R31
Altenhoff AM, Studer RA, Robinson-Rechavi M et al (2012) Resolving the ortholog conjecture: orthologs tend to be weakly, but significantly, more similar in function than paralogs. PLoS Comput Biol 8:e1002514
Altenhoff AM, Boeckmann B, Capella-Gutierrez S et al (2016) Standardized benchmarking in the quest for orthologs. Nat Methods 13:425–430
Studer RA, Robinson-Rechavi M (2009) How confident can we be that orthologs are similar, but paralogs differ? Trends Genet 25:210–216
Altenhoff AM, Dessimoz C (2009) Phylogenetic and functional assessment of orthologs inference projects and methods. PLoS Comput Biol 5:e1000262
Trachana K, Larsson TA, Powell S et al (2011) Orthology prediction methods: a quality assessment using curated protein families. BioEssays 33:769–780
Chen F, Mackey AJ, Vermunt JK et al (2007) Assessing performance of orthology detection strategies applied to eukaryotic genomes. PLoS One 2:e383
Dalquen DA, Altenhoff AM, Gonnet GH et al (2013) The impact of gene duplication, insertion, deletion, lateral gene transfer and sequencing error on orthology inference: a simulation study. PLoS One 8:e56925
Thomas PD, Campbell MJ, Kejariwal A et al (2003) PANTHER: a library of protein families and subfamilies indexed by function. Genome Res 13:2129–2141
Engelhardt BE, Jordan MI, Muratore KE et al (2005) Protein molecular function prediction by Bayesian phylogenomics. PLoS Comput Biol 1:e45
Cook SA (1971) The complexity of theorem-proving procedures. In: Proceedings of the third annual ACM symposium on theory of computing. ACM, New York, NY, pp 151–158
Sharan R, Ideker T (2006) Modeling cellular machinery through biological network comparison. Nat Biotechnol 24:427–433
Dewey CN, Pachter L (2006) Evolution at the nucleotide level: the problem of multiple whole-genome alignment. Hum Mol Genet 15 Spec No 1:R51–RR6
Górecki P (2004) Reconciliation problems for duplication, loss and horizontal gene transfer. In: Proceedings of the eighth annual international conference on research in computational molecular biology. ACM, New York, NY, pp 316–325
Hallett M, Lagergren J, Tofigh A (2004) Simultaneous identification of duplications and lateral transfers. In: Proceedings of the eighth annual international conference on Research in computational molecular biology. ACM, New York, NY, pp 347–356
Forslund K, Pereira C, Capella-Gutierrez S et al (2017) Gearing up to handle the mosaic nature of life in the quest for orthologs. Bioinformatics. https://doi.org/10.1093/bioinformatics/btx542
Guigó R, Muchnik I, Smith TF (1996) Reconstruction of ancient molecular phylogeny. Mol Phylogenet Evol 6:189–213
Bansal MS and Eulenstein O (2008) The multiple gene duplication problem revisited. Bioinformatics 24:i132–i13i138
Acknowledgments
We thank Stefan Zoller for helpful feedback on an earlier version of the manuscript. We gratefully acknowledge support by the Swiss National Science Foundation grant PP00P3_150654 to CD. Adrian M. Altenhoff and Natasha M. Glover contributed equally to this work.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2019 The Author(s)
About this protocol
Cite this protocol
Altenhoff, A.M., Glover, N.M., Dessimoz, C. (2019). Inferring Orthology and Paralogy. In: Anisimova, M. (eds) Evolutionary Genomics. Methods in Molecular Biology, vol 1910. Humana, New York, NY. https://doi.org/10.1007/978-1-4939-9074-0_5
Download citation
DOI: https://doi.org/10.1007/978-1-4939-9074-0_5
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-4939-9073-3
Online ISBN: 978-1-4939-9074-0
eBook Packages: Springer Protocols