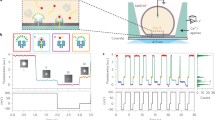
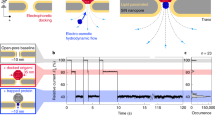
Abstract
Bacteriophage phi29 DNA packaging motor consists of a dodecameric portal channel protein complex termed connector that allows transportation of genomic dsDNA and a hexameric packaging RNA (pRNA) ring to gear the motor. The elegant design of the portal protein has facilitated its applications for real-time single-molecule detection of biopolymers and chemicals with high sensitivity and selectivity. The robust self-assembly property of the pRNA has enabled biophysical studies of the motor complex to determine the stoichiometry and structure/folding of the pRNA at single-molecule level. This chapter focuses on biophysical and analytical methods for studying the phi29 motor components at the single-molecule level, such as single channel conductance assays of membrane-embedded connectors; single molecule photobleaching (SMPB) assay for determining the stoichiometry of phi29 motor components; fluorescence resonance energy transfer (FRET) assay for determining the structure and folding of pRNA; atomic force microscopy (AFM) for imaging pRNA nanoparticles of various size, shape, and stoichiometry; and bright-field microscopy with magnetomechanical system for direct visualization of viral DNA packaging process. The phi29 system with explicit engineering capability has incredible potentials for diverse applications in nanotechnology and nanomedicine including, but not limited to, DNA sequencing, drug delivery to diseased cells, environmental surveillance, and early disease diagnosis.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Guo P (2002) Structure and function of phi29 hexameric RNA that drive viral DNA packaging motor: review. Prog Nucleic Acid Res Mol Biol 72:415–472
Aathavan K, Politzer AT, Kaplan A et al (2009) Substrate interactions and promiscuity in a viral DNA packaging motor. Nature 461:669–673
Rao VB, Feiss M (2008) The bacteriophage DNA packaging motor. Annu Rev Genet 42:647–681
Guo PX, Lee TJ (2007) Viral nanomotors for packaging of dsDNA and dsRNA. Mol Microbiol 64:886–903
Guo P, Grimes S, Anderson D (1986) A defined system for in vitro packaging of DNA-gp3 of the Bacillus subtilis bacteriophage phi29. Proc Natl Acad Sci U S A 83:3505–3509
Smith DE, Tans SJ, Smith SB et al (2001) The bacteriophage phi29 portal motor can package DNA against a large internal force. Nature 413:748–752
Simpson AA, Leiman PG, Tao Y et al (2001) Structure determination of the head-tail connector of bacteriophage phi29. Acta Crystallogr D57:1260–1269
Guasch A, Pous J, Ibarra B et al (2002) Detailed architecture of a DNA translocating machine: the high-resolution structure of the bacteriophage phi29 connector particle. J Mol Biol 315:663–676
Guo P, Erickson S, Anderson D (1987) A small viral RNA is required for in vitro packaging of bacteriophage phi29 DNA. Science 236:690–694
Guo P, Zhang C, Chen C et al (1998) Inter-RNA interaction of phage phi29 pRNA to form a hexameric complex for viral DNA transportation. Mol Cell 2:149–155
Shu D, Zhang H, Jin J et al (2007) Counting of six pRNAs of phi29 DNA-packaging motor with customized single molecule dual-view system. EMBO J 26:527–537
Zhang H, Endrizzi JA, Shu Y et al (2013) Crystal structure of 3WJ core revealing divalent ion-promoted thermostability and assembly of the phi29 hexameric motor pRNA. RNA 19:1226–1237
Guo P, Peterson C, Anderson D (1987) Prohead and DNA-gp3-dependent ATPase activity of the DNA packaging protein gp16 of bacteriophage phi29. J Mol Biol 197:229–236
Schwartz C, De Donatis GM, Fang H et al (2013) The ATPase of the phi29 DNA-packaging motor is a member of the hexameric AAA+ superfamily. Virology 443:20–27
Guo P, Noji H, Yengo CM et al (2016) Biological nanomotors with revolution, linear, or rotation motion mechanism. Microbiol Mol Biol Rev 80:161–186
Hendrix RW (1978) Symmetry mismatch and DNA packaging in large bacteriophages. Proc Natl Acad Sci U S A 75:4779–4783
Zhang H, Schwartz C, De Donatis GM et al (2012) “Push through one-way valve” Mechanism of Viral DNA Packaging. Adv Virus Res 83:415–465
Serwer P (2010) A hypothesis for bacteriophage DNA packaging motors. Virus 2:1821–1843
Feiss M, Rao VB (2012) The bacteriophage DNA packaging machine. Adv Exp Med Biol 726:489–509
Fang H, Jing P, Haque F et al (2012) Role of channel lysines and “push through a one-way valve” mechanism of viral DNA packaging Motor. Biophys J 102:127–135
Maluf NK, Feiss M (2006) Virus DNA translocation: progress towards a first ascent of mount pretty difficult. Mol Microbiol 61:1–4
Baumann RG, Mullaney J, Black LW (2006) Portal fusion protein constraints on function in DNA packaging of bacteriophage T4. Mol Microbiol 61:16–32
Hugel T, Michaelis J, Hetherington CL et al (2007) Experimental test of connector rotation during DNA packaging into bacteriophage phi29 capsids. PLoS Biol 5:558–567
Chang C, Zhang H, Shu D et al (2008) Bright-field analysis of phi29 DNA packaging motor using a magnetomechanical system. Appl Phys Lett 93:153902–153903
Schwartz C, De Donatis GM, Zhang H et al (2013) Revolution rather than rotation of AAA+ hexameric phi29 nanomotor for viral dsDNA packaging without coiling. Virology 443:28–39
Jing P, Haque F, Shu D et al (2010) One-way traffic of a viral motor channel for double-stranded DNA translocation. Nano Lett 10:3620–3627
Zhao Z, Khisamutdinov E, Schwartz C et al (2013) Mechanism of one-way traffic of hexameric phi29 DNA packaging motor with four electropositive relaying layers facilitating anti-parallel revolution. ACS Nano 7:4082–4092
De-Donatis G, Zhao Z, Wang S et al (2014) Finding of widespread viral and bacterial revolution dsDNA translocation motors distinct from rotation motors by channel chirality and size. Cell Biosci 4:30
Haque F, Li J, Wu H-C et al (2013) Solid-state and biological nanopore for real-time sensing of single chemical and sequencing of DNA. Nano Today 8:56–74
Branton D, Deamer DW, Marziali A et al (2008) The potential and challenges of nanopore sequencing. Nat Biotechnol 26:1146–1153
Venkatesan BM, Bashir R (2011) Nanopore sensors for nucleic acid analysis. Nat Nanotechnol 6:615–624
Healy K (2007) Nanopore-based single-molecule DNA analysis. Nanomedicine 2:459–481
Majd S, Yusko EC, Billeh YN et al (2010) Applications of biological pores in nanomedicine, sensing, and nanoelectronics. Curr Opin Biotechnol 21:439–476
Kasianowicz JJ, Robertson JW, Chan ER et al (2008) Nanoscopic porous sensors. Annu Rev Anal Chem (Palo Alto Calif) 1:737–766
Howorka S, Siwy Z (2009) Nanopore analytics: sensing of single molecules. Chem Soc Rev 38:2360–2384
Reiner JE, Balijepalli A, Robertson JW et al (2012) Disease detection and management via single nanopore-based sensors. Chem Rev 112:6431–6451
Wendell D, Jing P, Geng J et al (2009) Translocation of double-stranded DNA through membrane-adapted phi29 motor protein nanopores. Nat Nanotechnol 4:765–772
Jing P, Haque F, Vonderheide A et al (2010) Robust properties of membrane-embedded connector channel of bacterial virus Phi29 DNA packaging motor. Mol Biosyst 6:1844–1852
Geng J, Fang H, Haque F et al (2011) Three reversible and controllable discrete steps of channel gating of a viral DNA packaging motor. Biomaterials 32:8234–8242
Haque F, Geng J, Montemagno C et al (2013) Incorporation of viral DNA packaging motor channel in lipid bilayers for real-time, single-molecule sensing of chemicals and double-stranded DNA. Nat Protoc 8:373–392
Haque F, Wang S, Stites C et al (2015) Single pore translocation of folded, double-stranded, and tetra-stranded DNA through channel of bacteriophage Phi29 DNA packaging motor. Biomaterials 53:744–752
Geng J, Wang S, Fang H et al (2013) Channel size conversion of Phi29 DNA-packaging nanomotor for discrimination of single- and double-stranded nucleic acids. ACS Nano 7:3315–3323
Haque F, Lunn J, Fang H et al (2012) Real-time sensing and discrimination of single chemicals using the channel of Phi29 DNA packaging nanomotor. ACS Nano 6:3251–3261
Wang S, Haque F, Rychahou PG et al (2013) Engineered nanopore of Phi29 DNA-packaging motor for real-time detection of single colon cancer specific antibody in serum. ACS Nano 7:9814–9822
Cuervo A, Carrascosa JL (2012) Viral connectors for DNA encapsulation. Curr Opin Biotechnol 23:529
Shu D, Shu Y, Haque F et al (2011) Thermodynamically stable RNA three-way junctions for constructing multifuntional nanoparticles for delivery of therapeutics. Nat Nanotechnol 6:658–667
Haque F, Shu D, Shu Y et al (2012) Ultrastable synergistic tetravalent RNA nanoparticles for targeting to cancers. Nano Today 7:245–257
Shu D, Moll WD, Deng Z et al (2004) Bottom-up assembly of RNA arrays and superstructures as potential parts in nanotechnology. Nano Lett 4:1717–1723
Shu Y, Haque F, Shu D et al (2013) Fabrication of 14 Different RNA Nanoparticles for Specific Tumor Targeting without Accumulation in Normal Organs. RNA 19:766–777
Khisamutdinov EF, Jasinski DL, Guo P (2014) RNA as a boiling-resistant anionic polymer material to build robust structures with defined shape and stoichiometry. ACS Nano 8:4771–4781
Khisamutdinov E, Li H, Jasinski D et al (2014) Enhancing immunomodulation on innate immunity by shape transition among RNA triangle, square, and pentagon nanovehicles. Nucleic Acids Res 42:9996–10004
Jasinski D, Khisamutdinov EF, Lyubchenko YL et al (2014) Physicochemically tunable poly-functionalized RNA square architecture with fluorogenic and ribozymatic properties. ACS Nano 8:7620–7629
Lyubchenko YL, Shlyakhtenko LS, Ando T (2011) Imaging of nucleic acids with atomic force microscopy. Methods 54:274–283
Lyubchenko YL, Shlyakhtenko LS (2009) AFM for analysis of structure and dynamics of DNA and protein-DNA complexes. Methods 47:206–213
Lee SH, Shin JY, Lee A et al (2012) Counting single photoactivatable fluorescent molecules by photoactivated localization microscopy (PALM). Proc Natl Acad Sci U S A 109:17436–17441
Huang B, Wang WQ, Bates M et al (2008) Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science 319:810–813
Klar TA, Jakobs S, Dyba M et al (2000) Fluorescence microscopy with diffraction resolution barrier broken by stimulated emission. Proc Natl Acad Sci U S A 97:8206–8210
Annibale P, Vanni S, Scarselli M et al (2011) Quantitative photo activated localization microscopy: unraveling the effects of photoblinking. PLoS One 6:e22678
Lillemeier BF, Mortelmaier MA, Forstner MB et al (2010) TCR and Lat are expressed on separate protein islands on T cell membranes and concatenate during activation. Nat Immunol 11:90–96
Ori A, Banterle N, Iskar M et al (2013) Cell type-specific nuclear pores: a case in point for context-dependent stoichiometry of molecular machines. Mol Syst Biol 9:648
Sengupta P, Jovanovic-Talisman T, Lippincott-Schwartz J (2013) Quantifying spatial organization in point-localization superresolution images using pair correlation analysis. Nat Protoc 8:345–354
Zhang H, Shu D, Huang F et al (2007) Instrumentation and metrology for single RNA counting in biological complexes or nanoparticles by a single molecule dual-view system. RNA 13:1793–1802
Xiao F, Zhang H, Guo P (2008) Novel mechanism of hexamer ring assembly in protein/RNA interactions revealed by single molecule imaging. Nucleic Acids Res 36:6620–6632
Forster T (1946) Energiewanderung und fluoreszenz. Naturwissenschaften 6:166–175
Deniz AA, Dahan M, Grunwell JR et al (1999) Single-pair fluorescence resonance energy transfer on freely diffusing molecules: observation of Forster distance dependence and subpopulations. Proc Natl Acad Sci U S A 96:3670–3675
Norman DG, Grainger RJ, Uhrin D et al (2000) Location of cyanine-3 on double-stranded DNA: importance for fluorescence resonance energy transfer studies. Biochemistry 39:6317–6324
Iqbal A, Arslan S, Okumus B et al (2008) Orientation dependence in fluorescent energy transfer between Cy3 and Cy5 terminally attached to double-stranded nucleic acids. Proc Natl Acad Sci U S A 105:11176–11181
Chistol G, Liu S, Hetherington CL et al (2012) High degree of coordination and division of labor among subunits in a homomeric ring ATPase. Cell 151:1017–1028
Liu S, Chistol G, Hetherington CL et al (2014) A viral packaging motor varies its DNA rotation and step size to preserve subunit coordination as the capsid fills. Cell 157:702–713
Ibanez C, Garcia JA, Carrascosa JL et al (1984) Overproduction and purification of the connector protein of Bacillus subtilis phage f29. Nucleic Acids Res 12:2351–2365
Robinson MA, Wood JP, Capaldi SA et al (2006) Affinity of molecular interactions in the bacteriophage phi29 DNA packaging motor. Nucleic Acids Res 34:2698–2709
Guo Y, Blocker F, Xiao F et al (2005) Construction and 3-D computer modeling of connector arrays with tetragonal to decagonal transition induced by pRNA of phi29 DNA-packaging motor. J Nanosci Nanotechnol 5:856–863
Xiao F, Sun J, Coban O et al (2009) Fabrication of massive sheets of single layer patterned arrays using lipid directed reengineered phi29 motor dodecamer. ACS Nano 3:100–107
Cai Y, Xiao F, Guo P (2008) The effect of N- or C-terminal alterations of the connector of bacteriophage phi29 DNA packaging motor on procapsid assembly, pRNA binding, and DNA packaging. Nanomedicine 4:8–18
Ura Y, Beierle JM, Leman LJ et al (2009) Self-assembling sequence-adaptive peptide nucleic acids. Science 325:73–77
Shu Y, Shu D, Haque F et al (2013) Fabrication of pRNA nanoparticles to deliver therapeutic RNAs and bioactive compounds into tumor cells. Nat Protoc 8:1635–1659
Garver K, Guo P (2000) Mapping the inter-RNA interaction of phage phi29 by site-specific photoaffinity crosslinking. J Biol Chem 275(4):2817–2824
Huang LP, Guo P (2003) Use of PEG to acquire highly soluble DNA-packaging enzyme gp16 of bacterial virus phi29 for stoichiometry quantification. J Virol Methods 109:235–244
Lee CS, Guo P (1995) In vitro assembly of infectious virions of ds-DNA phage f29 from cloned gene products and synthetic nucleic acids. J Virol 69:5018–5023
Zhang H, Shu D, Wang W et al (2010) Design and application of single fluorophore dual-view imaging system containing both the objective- and prism-type TIRF. Proc SPIE 7571:757107–757108
Shu D, Zhang H, Petrenko R et al (2010) Dual-channel single-molecule fluorescence resonance energy transfer to establish distance parameters for RNA nanoparticles. ACS Nano 4:6843–6853
Sage D, Neumann FR, Hediger F et al (2005) Automatic tracking of individual fluorescence particles: application to the study of chromosome dynamics. IEEE Trans Image Process 14:1372–1383
Guo P (2014) Biophysical studies reveal new evidence for one-way revolution mechanism of bacteriophage phi29 DNA packaging motor. Biophys J 106:1837–1838
Acknowledgments
The research was supported by NIH grants U01-CA151648 and R01-EB012135. Funding to P.G.’s Endowed Chair in Nanobiotechnology position is by the William Fairish Endowment Fund. P.G. is a cofounder of Biomotor and Nucleic Acid Nanotechnology Development Corp., Ltd.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Haque, F., Zhang, H., Wang, S., Chang, CL., Savran, C., Guo, P. (2018). Methods for Single-Molecule Sensing and Detection Using Bacteriophage Phi29 DNA Packaging Motor. In: Lavelle, C. (eds) Molecular Motors. Methods in Molecular Biology, vol 1805. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-8556-2_21
Download citation
DOI: https://doi.org/10.1007/978-1-4939-8556-2_21
Published:
Publisher Name: Humana Press, New York, NY
Print ISBN: 978-1-4939-8554-8
Online ISBN: 978-1-4939-8556-2
eBook Packages: Springer Protocols