Abstract
When T cells scan the surface of antigen-presenting cells (APCs), they can detect the presence of just a few antigenic peptide/MHC complexes (pMHCs), in some cases even a single agonist pMHC. These are typically vastly outnumbered by structurally similar yet non-stimulatory endogenous pMHCs. How T cells achieve this enormous sensitivity and selectivity is still not clear, in particular in view of the rather moderate (1–100 μM) affinity that T-cell receptors (TCRs) typically exert for antigenic pMHCs. Experimental approaches that enable the control and quantification of physical input parameters within the context of the immunological synapse to precisely interrogate the molecular consequences of TCR-engagement, appear highly advantageous when searching for better answers.
We here describe the implementation of a biointerface that allows to experimentally define molecular distances between T-cell ligands as a means to correlate them with molecular dynamics of antigen engagement, downstream signaling, and the overall T-cell response. The basis of this biointerface is DNA origami nanostructures, which are (i) rigid and highly versatile platforms that can (ii) be embedded as laterally mobile entities within supported lipid bilayers and functionalized (iii) in a site-specific and orthogonal manner with (iv) one or more proteins of choice.
You have full access to this open access chapter, Download protocol PDF
Similar content being viewed by others
Key words
1 Introduction
When T cells scan the surface of antigen-presenting cells (APCs), they can detect the presence of just a few antigenic peptide/MHC complexes (pMHCs), in some cases even a single agonist pMHC. These are typically vastly outnumbered by structurally similar yet non-stimulatory endogenous pMHCs. How T cells achieve this enormous sensitivity and selectivity is still not clear, in particular in view of the rather moderate (1–100 μM) affinity that T-cell receptors (TCRs) typically exert for antigenic pMHCs. Experimental approaches, which leave the membranous environment and context provided by the living T-cell intact yet afford at the same time molecular or nanoscale resolution, appear highly advantageous when searching for better answers. This is why we and others have devised molecular imaging platforms, which involve planar glass-supported lipid bilayers (SLBs), which are functionalized with proteins of choice such as pMHCs, co-stimulatory, and adhesion molecules to serve as surrogate antigen-presenting cell for T cells. The use of SLBs for T-cell recognition offers a number of experimental advantages that make all the difference for arriving at meaningful conclusions: (i) the experimentalist exerts precise control over the nature and quantity of ligands the T cell gets to be exposed to; (ii) the planar nature of the setup reduces the imaging task from three to two dimensions, giving in turn rise to maximal spatiotemporal resolution; and (iii) images can be acquired in noise-attenuated total internal reflection (TIR) mode, which has so far remained a necessity for single molecule detection within the immunological synapse. Importantly, the stimulatory potency of SLB-resident pMHCs is very well preserved compared to cell surface-embedded pMHCs. Hence, while in principle artificial, SLBs are a good approximation of the physiological scenario a T cell encounters when approaching an APC. SLBs hence support molecular imaging, which is—unlike conventional biochemistry—per se noninvasive and yields molecular information in the highly dynamic context set by living T cells.
Despite the diffraction limit of visible light, a number of approaches have been successfully applied to extract via live cell imaging information at the nanoscopic level. These include (among others) tracking of single proteins with positional accuracies in the 10–50 nanometer range, high speed superresolution microscopy, and measurements of Förster resonance energy transfer (FRET) between fluorophore-labeled proteins of interest at the ensemble and single molecule level. The latter provides spatial information in a range of up to 8 nm. Collectively, these methodologies offer insights into “what happens” under chosen experimental settings (e.g., nature of TCR-pMHC pairing, antigen density, accessory molecules) within the confines of the immunological synapse with monitorable consequences for cellular physiology (e.g., downstream signaling, effector functions).
Highly complementary to molecular imaging is to experimentally define molecular distances between T-cell ligands as a means to correlate them with molecular dynamics of antigen engagement, downstream signaling, and the overall T-cell response. DNA origami nanostructures, the subject of this chapter, are (i) rigid and highly versatile platforms, which can (ii) be embedded as laterally mobile entities within supported lipid bilayers, and functionalized (iii) in a site-specific and orthogonal manner with (iv) one or more proteins of choice (Fig. 1). These qualities turn them into an exciting tool not only for T-cell biologists but also for the signaling, cell biological, and biophysical community, as the use of DNA origami combines nanoscale precision with a considerable degree of experimental flexibility and compatibility for anybody who wishes to dissect cell biological processes underlying cell-cell recognition. Here, we describe the use of SLB-embedded DNA origami nanostructures in the field of T-cell antigen recognition. However, we would like to emphasize that their application may also be of equal advantage in other areas of life sciences.
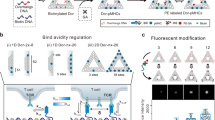
SLB-anchored DNA origami tiles for nanoscale ligand organization. (a) The production of the DNA origami-based biointerface involves the following steps: (1) folding of the DNA origami structure containing elongated staple strands at chosen sites and hybridization of biotinylated oligonucleotides, (2) attachment of divalent streptavidin (dSAv), (3) attachment of biotinylated pMHC, and (4) anchorage of DNA origami platforms to SLBs containing cholesterol-modified oligonucleotides. (b) Ligand-functionalized DNA origami tiles diffuse freely on the SLB. Tiles can be designed to harbor one, two, or more ligands. The platform size sets a minimum distance δ between ligands on adjacent platforms as they cluster in the course of T-cell activation. In the case of two or more ligands, distances d between ligands on the same tile can be pre-defined (we have used spacings of 10, 20, and 30 nm between two ligands)
In this chapter, we will provide recommendations for the production, protein-functionalization, purification, and SLB implementation of DNA origami structures for their use in live-cell recordings or immunofluorescence studies (Fig. 2). We will furthermore describe how to characterize such structures using single-molecule fluorescence microscopy.
2 Materials
2.1 Protein Purification
-
1.
Purified moth cytochrome c peptide with a GGSC linker (ANERADLIAYLKQATKGGSC).
-
2.
Purified moth cytochrome c peptide with a 3-amino-3-(2-nitrophenyl)propanoic acid (ANP) moiety (ANERADLIAYL[ANP]QATK).
-
3.
Escherichia coli BL-21 (DE3).
-
4.
IB detergent buffer (1% (w/v) deoxycholic acid, 1% (v/v) Triton X-100, 200 mM NaCl, 50 mM Tris–HCl pH 8.0 (4 °C), 1 mM EDTA).
-
5.
IB wash buffer A (1% (v/v) Triton X-100, 200 mM NaCl, 50 mM Tris–HCl pH 8.0 (4 °C), 1 mM EDTA).
-
6.
IB wash buffer B (200 mM NaCl, 50 mM Tris–HCl pH 8.0 (4 °C), 1 mM EDTA).
-
7.
Guanidine buffer (6 M guanidine hydrochloride, 50 mM Tris–HCl pH 8.0 (4 °C)).
-
8.
Resuspension buffer (25% sucrose, 50 mM Tris–HCl pH 8.0 (4 °C), 5 mM MgCl2).
-
9.
Amicon®Ultra centrifugal filters: 4 mL, 10 kDa.
-
10.
Amicon®Ultra centrifugal filters: 15 mL, 10 kDa.
-
11.
I-Ek refolding buffer (25% glycerol, 40 mM Na2HPO4, 10 mM NaH2PO4, 2 mM EDTA, 5 mM reduced glutathione, 0.5 mM oxidized glutathione, 0.1 mM PMSF (dissolved in isopropanol, 100 mM Stock)).
-
12.
BirA biotin ligase (Avidity).
-
13.
Biomix A (0.5 M bicine buffer pH 8.3).
-
14.
Biomix B (100 mM ATP, 100 mM magnesium acetate, 0.5 mM d-biotin).
-
15.
10× biotin (0.5 mM d-biotin).
-
16.
2 M potassium glutamate.
-
17.
Glutathione agarose.
-
18.
1 M HEPES pH 7.0.
-
19.
100 mM Phenylmethylsulfonylfluorid (PMSF) dissolved in isopropanol.
-
20.
pET21a(+) streptavidin dead.
-
21.
pET21a(+) streptavidin alive hexa-glutamate tag.
-
22.
pET21a(+) I-Ek α AVItag subunit.
-
23.
pET28a(+) I-Ek β subunit.
-
24.
ÄKTA pure (Cytiva) or any other protein purification system.
-
25.
Mono Q™ 5/50 GL (Cytiva).
-
26.
Superdex™ 75 10/300 GL (Cytiva).
-
27.
Superdex™ 200 10/300 GL (Cytiva).
-
28.
14.4.4S mAb cross-linked to cyanogen bromide-activated agarose (Merck).
-
29.
Analytical HPLC system (Agilent).
-
30.
MALDI-TOF mass spectrometry system (e.g., rapifleX, Bruker).
-
31.
nanoHPLC-nanoESI-Linear-Trap-Quadrupole-Orbitrap mass spectrometry system (e.g. UltiMate 3000 RSLCnano, LTQ Orbitrap Velos, Dionex/Thermo Fisher Scientific).
-
32.
Ultra-purity silica-based C18 HPLC column (e.g., Pursuit XRs C18, 5 μm, 250 × 21.2 mm).
-
33.
Immobilized TCEP disulfide reducing gel.
-
34.
Acetonitrile (HPLC-grade).
-
35.
Trifluoroacetic acid.
-
36.
LB agar medium.
-
37.
LB medium.
-
38.
100 mg/mL Ampicillin in ddH2O.
-
39.
50 mg/mL Kanamycin in ddH2O.
-
40.
1 M Isopropyl-b-D-thiogalactopyranoside (IPTG) in ddH2O.
-
41.
Lyophilized DNase I.
-
42.
Lyophilized lysozyme.
-
43.
Deoxycholic acid, powder.
-
44.
Triton X-100, solution.
-
45.
Guanidine hydrochloride, powder.
-
46.
0.22 μm and 0.45 μm syringe filters.
-
47.
0.22 μm centrifuge tube filters (Spin-X, Corning).
-
48.
Glycerol, solution.
-
49.
Oxidized glutathione.
-
50.
Reduced glutathione.
-
51.
CAPS elution buffer (50 mM CAPS, 10% glycerol, 0.15 M NaCl, 2 mM EDTA, adjusted with NaOH to pH 11.5).
-
52.
Neutralization buffer (600 mM Tris–HCl pH 8.0, 0.15 mM NaCl).
-
53.
10% (w/v) NaN3.
-
54.
Citric acid buffer pH 4.9.
-
55.
Pressure-based stirred cell with ultrafiltration cellulose membrane disc (10 kDa).
2.2 Microscopy Setup
A microscope setup with TIR illumination and single-molecule detection capabilities is required. Critical elements are as follows: an inverted microscope setup with TIR option and TIR objective (numerical aperture ≥1.45), a fast EM-CCD camera, diode, or solid-state lasers with 488 nm, 532 nm, and 640 nm wavelengths that allow modulation of laser intensities as well as timings in the millisecond range.
2.3 Other Components
-
1.
Coverslips: 24 × 60 mm #1.5, borosilicate, for example, Menzel-Gläser, Thermo Fisher or equivalent.
-
2.
PCR tube.
-
3.
M13mp18 DNA scaffold (Tilibit or equivalent).
-
4.
50 μM DNA staple strands in TE buffer at −20 °C.
-
5.
1 M Tris–HCl, pH 8.0, solution.
-
6.
5 M NaCl solution.
-
7.
0.5 M EDTA solution adjusted to pH 8.0 with NaOH.
-
8.
1 M MgCl2 solution.
-
9.
TE buffer: 10 mM Tris–HCl, pH 8.0, 1 mM EDTA.
-
10.
10× Folding buffer (FoB): 50 mM Tris–HCl, pH 8.0, 500 mM NaCl, 10 mM EDTA.
-
11.
Purification buffer (PuB): 5 mM Tris–HCl, pH 8.0, 50 mM NaCl, 1 mM EDTA, 5 mM MgCl2.
-
12.
Sticky-Slide 8-Well chamber (Ibidi or equivalent).
-
13.
ddH2O.
-
14.
DNA LoBind™tube (Eppendorf or equivalent) for DNA origami preparation.
-
15.
Amicon®Ultra centrifugal filters: 0.5 mL, 100 kDa.
-
16.
Biopur®tubes (Eppendorf or equivalent) for DNA origami storage.
-
17.
Amicon®Ultra centrifugal filters: 2.0 mL, 100 kDa.
-
18.
Gel Loading Dye (6×), no SDS.
-
19.
TAE buffer: 40 mM Tris–HCl base, 10 mM acetic acid, 1 mM EDTA pH 8.3.
-
20.
SYBR™Gold nucleic acid stain.
-
21.
YOYO™-1 Iodide (Thermo Scientific or equivalent), a DNA stain for two-color colocalization.
-
22.
1 kB DNA ladder.
-
23.
Hank’s Balanced Salt Solution (HBSS): 140 mM NaCl, 5 mM KCl, 0.3 mM Na2HPO4, 0.4 mM KH2PO4, 6 mM D-glucose, 4 mM NaHCO3.
-
24.
Imaging buffer: HBSS +1 mM CaCl2, 0.4 mM MgSO4, 0.5 mM MgCl2.
-
25.
UV spectrophotometer.
-
26.
10× PBS: 1.37 M NaCl, 27 mM KCl, 80 mM Na2HPO4, 20 mM KH2PO4, and 3 mM KCl, pH 7.4.
-
27.
1× PBS: 10× PBS diluted with ddH2O.
-
28.
5 mg/mL 1,2-dioleoyl-sn-glycero-3-[(N-(5-amino-1-carboxypentyl)iminodiacetic acid)succinyl] (nickel salt) (DGS Ni-NTA) in chloroform.
-
29.
10 mg/mL 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine (POPC) in chloroform.
-
30.
Bath sonicator.
-
31.
Plasma chamber, also referred to as plasma cleaner.
-
32.
Fiolax™ Borosilicate test tube or equivalent.
-
33.
Biotin-DNA (Biomers) (BT-TEG-5’-ACATGACACTACTCCAC-3′).
-
34.
TetraSpeck™ microspheres, 0.1 μm, fluorescent blue/green/orange/dark red Thermo Fisher.
-
35.
Alexa Fluor®555 C2 maleimide.
-
36.
Abberior Star 635P, maleimide.
-
37.
Cholesterol-DNA (5’-GGCTAAATATGCTAGGACTCT-3′-TEG-cholesterol).
-
38.
10% Bovine serum albumin in ddH2O.
-
39.
Recombinant ICAM-1 extracellular domain with C-terminal 10xHis-tag.
-
40.
Recombinant B7-1 extracellular domain with C-terminal 10xHis-tag.
3 Methods
3.1 Expression of Streptavidin and I-Ek Subunits as Insoluble Inclusion Bodies in E. coli
-
1.
For expression of the desired protein, transform E. coli BL21 (DE3) with pET expression vectors encoding for (i) the alive/dead streptavidin subunits or (ii) I-Ek α-AVItag / I-Ek β and plate transformed bacteria onto LB agar dishes supplemented with 100 μg/mL ampicillin (pET21a+) or 50 μg/mL kanamycin (pET28a+). Incubate overnight at 37 °C.
-
2.
Pick a single colony and start overnight culture in LB medium supplemented with either 100 μg/mL ampicillin or 50 μg/mL kanamycin. Prepare 10 mL of overnight culture per 1 l of expression culture and shake at 180–200 rpm overnight at 37 °C. We recommend 2 l expression culture per streptavidin subunit and 6 l per I-Ek subunit when using LB medium supplemented with antibiotics.
-
3.
Dilute overnight culture 1:100 in 0.5–1.0 l LB medium supplemented with 100 μg/mL ampicillin or 50 μg/mL kanamycin. Use a culture flask with a total volume of 3 l for expression and incubate at 37 °C while shaking at 180–220 rpm. Monitor growth of bacteria with a spectrophotometer until OD600 = 0.5 (~2 h). Optionally, take a pre-induction sample (1 mL), centrifuge at 15,000 g for 1 min in a table top micro centrifuge (e.g., Eppendorf 5430) and freeze the pellet at −20 °C for later verification of protein induction via SDS-PAGE analysis.
-
4.
Induce protein expression by adding isopropyl-b-D-thiogalactopyranoside (IPTG) at a concentration of 1 mM. Shake expression culture at 180–220 rpm and 37 °C for 4–5 h.
-
5.
Harvest bacteria. Centrifuge culture at 8000 g for 15–20 min at 4 °C in 1 l containers. Collect pellet and dissolve in 10 mL resuspension buffer per liter culture. Transfer to 50 mL tubes (maximal volume 30 mL), add DNase I at 30 μg/mL and lysozyme at 1 mg/mL and sonicate at room temperature 3–5 rounds using a tip sonicator, each time for 1 min. Optionally, for later verification of protein induction via SDS-PAGE, take a post-induction aliquot (1 mL) prior to harvesting, centrifuge at 15,000 g for 1 min in a table top micro centrifuge and freeze at −20 °C.
-
6.
After sonication, centrifuge for 10 min at 10,000 g, collect the pellet, and resuspend in 10–20 mL of IB detergent buffer (4 °C). Use a spatula to resuspend inclusion bodies in the wash buffer. Centrifuge for 10 min at 10,000 g and repeat the solubilization and centrifugation step once.
-
7.
Collect inclusion bodies and wash with 10–20 mL IB wash buffer A. Centrifuge for 10 min at 10,000 g and repeat the washing and centrifugation step five times.
-
8.
Collect inclusion bodies and wash with 10–20 mL IB wash buffer B. Centrifuge for 10 min at 10,000 g and repeat the washing and centrifugation step once. Purified inclusion bodies appear chalky and can be frozen and stored at ≤ −20 °C.
-
9.
Solubilize inclusion bodies in guanidine buffer (4 °C) at a concentration of 10–20 mg/mL. Estimate protein concentration by measuring the OD280 of 1:50 dilutions using a spectrophotometer. Blank against guanidine buffer. Filter solution through a 0.45 μm syringe filter and store the solubilized protein at ≤ −20 °C.
-
Streptavidin subunit alive hexa-glutamate tag: MW = 14,177, ε = 41,940.
-
Streptavidin subunit dead: MW = 13,372, ε = 41,940.
-
I-Ek α (AVI-tag) subunit: MW = 23,806, ε = 31,970.
-
I-Ek β subunit: MW = 23,191, ε = 39,420.
-
3.2 Refolding and Purification of Trans dSav
Divalent streptavidin (dSAv) is a heterotetrametric protein consisting of two dead subunits (not capable of biotin binding) and two alive subunits (capable of biotin binding). The two alive subunits can either point both in the same direction (cis configuration) or point to opposite directions (trans configuration) [1, 2]. To produce streptavidin tetramers with controlled divalent pairing of alive and dead subunits in the trans divalent configuration, we extended the alive streptavidin subunit C-terminally with a hexa-glutamate tag to enable purification via ion-exchange chromatography as described in [1] .
-
1.
Express and purify inclusion bodies of alive and dead streptavidin subunits as described above.
-
2.
Solubilize inclusion bodies of each streptavidin subunit at a concentration of 10 mg/mL in guanidine buffer (4 °C) and filter through a 0.45 μm syringe filter.
-
3.
Premix alive and dead subunits in a 1:1 or 2:1 molar ratio to maximize the refolding yield of dSav. For example, premix 5.3 mg alive with 5 mg dead subunits to reach a 1:1 molar ratio in the final refolding reaction (1.5 μM of each subunit in 250 mL 1× PBS).
-
4.
Precool 250 mL 1× PBS to 4 °C in a 500 mL glass beaker in the cold room. Add a stir bar and stir the PBS rapidly to create a vortex. Add the sav mixture dropwise (using a 200 μL pipette) to the stirring PBS to avoid protein aggregates. The refolding reaction is most successful when the solution remains clear. Continue slow stirring overnight at 4 °C.
-
5.
After refolding, concentrate the streptavidin tetramer mixture to 10 mL using a pressure-based ultrafiltration stirred cell equipped with a 10 kDa cutoff cellulose membrane. Further concentration and buffer exchange to 20 mM Tris–HCl pH 8.0 (4 °C) can be achieved with Amicon®Ultra:15 centrifugal filters (10 kDa) and dialysis. Of note, pre-rinse Amicon®Ultra centrifugal filters with buffer prior to applying the protein solution. Centrifuge the filters with 5000 g at 4 °C until the desired volume or concentration is reached. Alternatively, precipitate streptavidin from the refolding mixture using ammonium sulfate as described in [1, 2] or enrich the properly folded streptavidin using an iminobiotin-Sepharose affinity column as described in [1].
-
6.
After buffer exchange to 20 mM Tris–HCl pH 8.0 (4 °C), load the refolded streptavidin mixture onto an ion exchange chromatography column (MonoQ 5/50 GL) with a flow-rate of 1 mL/min using an ÄKTA pure protein purification system. The tetrameric trans divalent streptavidin can be separated from dead, monovalent, cis divalent, trivalent, or tetravalent streptavidin based on the hexa-glutamate tag located on the alive subunit. After loading, wash the MonoQ column with loading buffer (4 °C) and 0.1 M NaCl for 5–10 mL to elute dead streptavidin, before applying a linear gradient from 0.2–0.35 M NaCl over 90 mL. Collect 1 mL fractions with a flow rate of 1 mL/min [1].
-
7.
Concentrate fractions containing trans dSav with Amicon®Ultra:4 centrifugal filters (10 kDa) and purify the protein via S200 gel filtration (Superdex 200 10/300) using an ÄKTA pure protein purification system. Collect monomeric trans dSav, concentrate to 0.2–1 mg/mL, supplement with 50% glycerol, and store at −20 °C.
3.3 Refolding I-Ek in Complex with a Placeholder Peptide
The murine MHC molecule I-Ek can be refolded in vitro as heterotrimeric protein complex composed of a polypeptide chain, the I-Ek α subunit and the I-Ek β subunit. The I-Ek α subunit is C-terminally extended with a BirA biotin ligase recognition site, while the β subunit is devoid of a tag.
-
1.
Express and purify inclusion bodies of the I-Ek α subunit (AVI-tag) and the I-Ek β subunit (no tag) as described above. Solubilize 200 mg inclusion bodies of each subunit at a concentration of 20 mg/mL in guanidine buffer (4 °C) and filter through a 0.45 μm syringe filter.
-
2.
Prepare 4 l of I-Ek refolding buffer, check pH (pH 7–8) and transfer to the cold room.
-
3.
Dissolve 74.7 mg of (preferably HPLC-purified) placeholder peptide (ANERADLIAYL[ANP]QATK) in 20 mL ddH20 and add to the I-Ek refolding buffer (final concentration of ANP peptide is 10 μM).
-
4.
Stir refolding buffer vigorously to form a vortex. Add 190.4 mg I-Ek α (final concentration 2 μM) and 185.5 mg I-Ek β (final concentration 2 μM) dropwise to the refolding buffer using a 200 μL pipette.
-
5.
Incubate the I-Ek refold for at least 3 days in the cold room with slow stirring.
3.4 Purification of I-Ek/ANP
-
1.
After completion of the I-Ek refolding reaction, centrifuge the solution at >15,000 g for >30 min at 4 °C. Collect the supernatant, filter through 0.45 μm syringe filters and add 0.1 mM PMSF.
-
2.
Load supernatant onto a custom-prepared 14.4.4S monoclonal antibody (mAb) affinity column (2 mL bed volume) with a flowrate of ~1 mL/min. Wash column with 10 column volumes of 1× PBS and elute I-Ek/ANP in 1 mL fractions with CAPS elution buffer. Prior to elution, prepare 0.5 mL of neutralization buffer in 1.5 mL tubes to immediately neutralize the high pH of the CAPS elution buffer. After elution, wash the 14.4.4S mAb column with 1 × PBS and store in 1× PBS supplemented with 0.05% NaN3 at 4 °C. Optionally, load the eluted and refolded I-Ek a second time onto the 14.4.4S mAb affinity column.
-
3.
Subject the eluted protein to S200 gel filtration (Superdex 200 10/300) to remove aggregates and concentrate fractions containing monomeric I-Ek/ANP with Amicon®Ultra:4 centrifugal filters (10 kDa) to 1–2 mg/mL. Snap freeze aliquots in liquid N2 for storage at −80 °C or directly proceed with biotinylation.
3.5 Site-Specific Biotinylation of I-Ek/ANP Using the BirA Biotin Ligase
For site-specific biotinylation of I-Ek/ANP with the GST-tagged BirA biotin ligase, we refer to the protocol described by Avidity (Avitag™ Technology).
-
1.
Prepare 1–2 mg I-Ek/ANP at a concentration of 20–40 μM in 1× PBS. Prepare 10× stocks of Biomix A, Biomix B and additional biotin (0.5 mM d-biotin).
-
2.
Dilute I-Ek/ANP (in 1× PBS) at least two- to threefold with ddH20 to reduce the NaCl concentration in the biotinylation reaction as NaCl strongly reduces the activity of the BirA biotin ligase. For example, mix 100 μL Biomix A (10×), 100 μL Biomix B (10×), 100 μL biotin (10×), 300 μL ddH20, and 300 μL of I-Ek/ANP protein solution. To ensure rapid biotinylation, increase the substrate concentration again to 20–40 μM using Amicon®Ultra:4 centrifugal filters (10 kDa). Finally, add at least 1 μg BirA ligase per 4 nmol substrate and incubate at 30 °C for 40 min. To reach quantitative biotinylation, adjust the amount of BirA biotin ligase to the quantity and concentration of the substrate as described by the manufacturer Avidity. In case of incomplete biotinylation of the substrate, increase the amount of BirA biotin ligase or the concentration of the substrate. If low protein yields after biotinylation occur, substitute NaCl for 0.2 M potassium glutamate and biotinylate overnight at 4 °C.
-
3.
After biotinylation, supplement the reaction mix with 100 mM NaCl and remove the BirA ligase using glutathione agarose according to the manufacturer’s instructions. Incubate the sample at 4 °C for 1 h, centrifuge at 1000 g for 1 min to remove the resin and filter the supernatant through a 0.22 μm centrifuge tube filter (spin at 10,000 g for 2 min).
-
4.
Purify the biotinylated I-Ek/ANP via S200 gel filtration (Superdex 200 10/300), collect monomeric fractions and concentrate the protein to ~1 mg/mL using Amicon®Ultra:4 centrifugal filters (10 kDa). Snap freeze biotinylated I-Ek/ANP with liquid N2 for storage at −80 °C or directly proceed with peptide exchange.
-
5.
Verify quantitative biotinylation of I-Ek/ANP with a streptavidin-based gel shift assay and SDS-PAGE analysis.
3.6 Site-Specific Labeling of a Peptide with Maleimide-Conjugated Dyes
For site-specific labeling of peptides presented by I-Ek, we typically extend suitable peptides (e.g., moth cytochrome c peptide, ANERADLIAYLKQATK) with a GGSC-linker at the C-terminus (MCC-GGSC), which allows for maleimide-based conjugation to fluorophores as described in (Huppa et al. 2010).
-
1.
Solubilize peptides in ddH20 or any other appropriate buffer and purify via reversed-phase HPLC (e.g., Pursuit XRs C18 5 μm 250 × 21.2 mm column) using an 0–100% linear gradient from an ionic solvent (0.1% trifluoroacetic acid in ddH20) to an organic solvent mix (0.1% trifluoroacetic acid, 10% ddH20, 89.9% acetonitrile) over 100 min with a flowrate of 5 mL/min. Collect peak fractions.
-
2.
Verify peak fractions via mass spectrometry (e.g., MALDI-TOF or nanoHPLC-nanoESI Orbitrap) and lyophilize peptide fractions with the correct molecular mass. Store lyophilized peptides at ≤ −20 °C.
-
3.
Solubilize peptides at a concentration of 1–5 mg/mL (0.5–2.5 mM) in 1× PBS or any other buffer at pH 6.5–7.5 recommended for maleimide-based conjugation (e.g., 10–100 mM Tris–HCl or HEPES).
-
4.
Reduce oxidized sulfhydryl groups with immobilized TCEP (Tris[2-carboxyethyl] phosphine hydrochloride) disulfide reducing gel according to the manufacturer’s instructions. Add one volume of TCEP resin (washed once with 1× PBS) to one or two volumes of peptide solution and incubate for 1 h at room temperature. After incubation, centrifuge the sample 1000 g for 1 min and filter the supernatant through a 0.22 μm centrifuge tube filter (spin at 10,000 g for 2 min).
-
5.
Dissolve maleimide-conjugated dyes (e.g., Alexa Fluor™ 555 C2 Maleimide) at a concentration of 10–20 mM in DMSO or DMF immediately before use. Add sufficient maleimide-conjugated dyes from the stock solution to give approximately 2 moles of dye for each mole of peptide and incubate for 2 h at room temperature or overnight at 4 °C. We recommend mixing at least 300 μg MCC-GGSC with 356 μg Alexa Fluor™ 555 C2 Maleimide (1:2 molar ratio) to obtain sufficient amounts of dye-conjugated peptide for a quantitative I-Ek peptide exchange. The reaction can be stopped with an excess of glutathione.
-
6.
Purify the reaction mix via reversed-phase HPLC to separate dye-conjugated peptide from unconjugated peptide or dye as described above. Verify peak fractions with mass spectrometry and lyophilize the site-specifically labeled peptide for storage at −80 °C.
3.7 Exchange of the I-Ek-Associated ANP Placeholder Peptide with Site-Specifically Labeled Peptides
The ANP placeholder peptide can be substituted with any peptide that binds into the peptide-binding cleft of I-Ek under slightly acidic conditions (pH 5.1) as described in [3, 4].
-
1.
Dilute 0.25 mg of I-Ek/ANP (~1 mg/mL) in 1× PBS to a final concentration 20 μM. Dissolve the lyophilized and site-specifically labeled peptide (e.g., MCC-AF555) in 25–50 μL 1× PBS and add the solubilized peptide to I-Ek/ANP. Mix carefully.
-
2.
For quantitative peptide exchange, add citric acid buffer pH 4.9 at a final concentration of 200 mM to the peptide exchange reaction to reach an exact pH of 5.1. Incubate for 1 h at room temperature and subsequently centrifuge for 2 min at 15,000 g to pellet and remove denatured protein. Collect supernatant and incubate for 1–3 days at room temperature.
-
3.
Subject the protein-peptide solution to an S75 or S200 gel filtration step (e.g., Superdex 75 10/300 or Superdex 200 10/300) to remove protein aggregates and unbound peptide. Concentrate fractions containing monomeric I-Ek/MCC-AF555 with Amicon®Ultra:4 centrifugal filters (10 kDa) and store the protein at a concentration of 0.2–1 mg/mL in 1× PBS supplemented with 50% glycerol at −20 °C.
-
4.
Determine the protein-to-dye ratio and consequently the efficiency of peptide exchange by measuring the absorption at 280 nm and 555 nm with a spectrophotometer. A protein-to-dye ratio of 1 reflects quantitative peptide loading.
3.8 DNA Origami Preparation
DNA origami structures are typically assembled from a long, single-stranded scaffold DNA and short, specifically designed staple strands in a one-pot folding reaction using a thermal cycler. We have characterized in detail three rectangular, single-layer DNA origami tiles of three different sizes (30 × 20 nm, 65 × 54 nm, 100 × 70 nm) [5] based on the M13mp18 scaffold [6]. In these core structures, specific staple strands can be elongated for functionalization with ligands (on the top side of the tile) or for anchorage to the SLB via hybridization to cholesterol-modified DNA oligonucleotides (on the bottom side). For functionalization, we either elongate one centrally located staple strand or two strands at 10, 20, or 30 nm distance; at the bottom side, the DNA origami feature six to ten elongations for the attachment of cholesterol-DNA (Fig. 1). Layouts can easily be adapted for individual purposes using the open-source software caDNAno [7] (see Note 1).
-
1.
Mix DNA (prewarmed to 24 °C), MgCl2, and folding buffer FoB 10× (50 mM Tris–HCl (pH = 8.0), 500 mM NaCl, 10 mM EDTA) in a DNA LoBind® tube (see Note 2). A representative folding protocol for 100 × 70 nm DNA origami tiles is shown in table below. Modified (i.e., biotinylated and/or fluorescently labeled) oligonucleotides are designed to be complementary in sequence to elongated staples for ligand attachment on the top side (“V sequence” 5′-ACATGACACTACTCCAC-3′), and cholesterol- DNA is complementary in sequence to elongations on the bottom side (“Z sequence” (5′-GGCTAAATATGCTAGGACTCT-3′)) (see [8]).
Component | Concentration | DNA strands [n] | Volume [μL] | Final concentration |
|---|---|---|---|---|
M13mp18 scaffold | 100 nM | – | 10.0 | 10 nM |
Folding buffer (FoB) | 10× | – | 10.0 | 1× |
MgCl2 | 100 mM | – | 12.5 | 12.5 mM |
Master mix (MM) | 50 μM | 175 | 34.8 | 100 nM/staple |
Surface attachment mix (SAM) | 50 μM | 8 | 1.6 | 100 nM/staple |
Ligand attachment mix (LAM) | 50 μM | 1 | 0.2 | 100 nM/staple |
Biotin-DNA (V-sequence) | 50 μM | 1 | 1.0 | 500 nM/staple |
H2O, ultrapure | 31.5 | |||
Total volume | 100.0 |
-
2.
Distribute the folding mix to ten PCR tubes and place them into a thermal cycler (CFX Connect Real-Time PCR Detection System) following a thermal protocol optimized for the structure to be folded. At this stage, DNA origami can be stored in the freezer at −20 °C for up to 3 months. The optimized thermal protocol for the described DNA origami layouts is (24 °C–90 °C, 10 °C min-1; 90 °C, 15 min; 90 °C–4 °C, 1 °C min-1; 4 °C, 6 h). The critical temperature for DNA duplex formation is between 55 °C and 65 °C depending on the GC content.
3.9 DNA Origami Purification
Purification of DNA origami structures can be achieved via several methods [9], with yields and purity depending on the specific structure. For simple structures such as the described tiles, spin column purification (with, e.g., Amicon®Ultra centrifugal filters) constitutes an adequate and user-friendly option. For more complex DNA nanoarchitectures (e.g., 3D tripods), we recommend purification via agarose gel electrophoresis or FPLC to remove larger DNA aggregates.
-
1.
Prepare 5 mL aliquots of the PuB. The lower Mg2+ content (5 mM) of the PuB compared to the folding mix improves recovery yields of correctly folded DNA origami structures from filter membranes. Always use freshly prepared PuB.
-
2.
Pre-rinse Amicon®Ultra:0.5 (100 kDa cutoff membrane) with PuB according to the manufacturer’s instructions by evenly distributing 500 μL of PuB on the filter (do not touch the filter) followed by centrifugation at 5000 g for 5 min at 24 °C. Discard the flow-through and insert the filter into a clean tube. The molecular weight of staple strand DNA is ≤10 kDa, but in our hands, 100 kDa membranes yield better separation.
-
3.
Mix 100 μL DNA origami solution with 400 μL PuB and evenly distribute the solution on the filter. Centrifuge at 7000 g for 5 min at 24 °C and discard the flow-through.
-
4.
Add PuB to a volume of 500 μL to the filter followed by spinning at 7000 g for 5 min at 24 °C. Discard the flow-through and repeat the procedure one more time. If the volume in the filter exceeds 50 μL, spin five to ten more minutes at 7000 g at 24 °C.
-
5.
Add 50 μL PuB to a new clean collection tube, invert the filter and put it into the collection tube for recovering purified DNA origami structures. Spin at 5000 g for 4 min at 24 °C. Adjust the concentration of DNA origami to 10 nM by adding appropriate amounts of PuB. At this stage, DNA origami structures can be stored in Biopur® tubes for up to 4 weeks at −20 °C.
3.10 DNA Origami Functionalization Strategy Using Divalent Streptavidin
We have recently assessed multiple different strategies for the site-specific decoration of DNA origami structures with regard to achieving optimal yields and full functionality of attached proteins [10]. We here describe a functionalization strategy based on dSA. In principle, also commercially available tetravalent streptavidin can be used, but this will likely lead to structures featuring up to three pMHC molecules per functionalization site [10].
-
1.
Add 50 μL of a 10 nM DNA origami solution into a DNA LoBind® tube, prewarm to 24 °C and add a 10× molar excess of dSAv. Incubate for 30 min at 24 °C.
-
2.
Pre-incubate an Amicon®Ultra:2.0 centrifugal filter (100 kDa) with 2 mL PuB. Spin at 4000 g for 10 min at 4 °C. Discard the flow-through and reinsert the filter into the tube.
-
3.
Mix the DNA origami solution with PuB to a final volume of 1.5 mL and evenly distribute the solution on the filter. Centrifuge at 4000 g for 15 min at 4 °C and discard the flow-through. Add 1.5 mL PuB to the filter followed by spinning at 4000 g for 15 min at 4 °C. Discard the flow-through. If the volume in the filter exceeds 50 μL, spin 5–10 min at 4000 g at 4 °C.
-
4.
Invert the filter, add 50 μL PuB to the conical collection tube and spin at 2000 g for 4 min at 4 °C. Adjust the concentration of DNA origami to 5 nM with PuB.
-
5.
Incubate the desired amount of DNA origami (pre-warmed to 24 °C) at 10× molar excess of AF555-conjugated and site-specifically biotinylated pMHC in a DNA LoBind® tube for 60 min at 24 °C.
-
6.
Repeat steps 2–4.
-
7.
Functionalized DNA origami structures can be stored at this step (or after step 4) in Biopur® tubes up to 1 week at 4 °C.
3.11 DNA Origami Quality Control: Gel Electrophoresis
We recommend verifying the integrity of DNA origami structures as well as the successful functionalization applying several different methods including gel electrophoresis, high-speed atomic force microscopy (hs-AFM), electron microscopy, DNA Points Accumulation for Imaging in Nanoscale Topography (DNA-PAINT), and others. Representative images are shown in Fig. 3. We here describe the quality control via gel electrophoresis as a rapid initial quality feedback, which is readily available in most biochemical labs. We recommend performing gel electrophoresis after folding, after initial purification, and after dSAv attachment. hs-AFM imaging and DNA-PAINT imaging are recommended after dSAv functionalization but before addition of biotinylated pMHC.
-
1.
Prepare a 1% w/v agarose gel with 1× Tris-acetate-EDTA (TAE) buffer pH 8; stain with, for example, Sybr™-Gold. For larger or smaller DNA origami structures, agarose content can be varied between 0.5% and 2.0%. Load ~5 ng DNA origami, the M13mp18 scaffold and a 1kB DNA ladder on the gel and run the gel for 75 min at 100 V and 24 °C in 1× TAE pH 8 supplemented with 10 mM MgCl2. Avoid currents above 150 mA. The presence of MgCl2 is necessary to maintain DNA origami integrity but may result in temperature peaks (>>40 °C), which may damage the DNA origami as well as attached proteins. We recommend to regularly monitor the temperature and, if necessary, run the gel on ice.
-
2.
Image the gel with an UV-light source. Correctly folded DNA origami will appear as a discrete band shifted to a larger apparent size when compared to the M13mp18 scaffold.
DNA origami quality control. (a) Agarose gel electrophoresis of DNA origami platforms. Schematic sketches above the individual lanes indicate the different DNA origami layouts functionalized with dSAv. from left to right: M13mp18 scaffold, S, M, L DNA origami. (b) Schematics of a 100 × 70 nm (L) DNA origami tile. Circles indicate available modification sites on the top side of the DNA origami tile; full circles indicate sites used for functionalization with two ligands at 20 nm distance (Ldiv 20 nm). The distances are approximated with 0.34 nm per base pair along the helical axis and with 2 nm per helix perpendicular to the helical axis [6]. Interhelical gaps were assigned 1 nm, based on the spacing of crossovers between helices. Distances are given in nm. (c) hs-AFM image of an L DNA origami platform featuring two dSAvs spaced 20 nm apart. Scale bar, 25 nm. (d) Mapping of dSAv positions on DNA origami platforms via DNA-PAINT. (i) Biotinylated ligands are replaced with biotinylated DNA-PAINT docking strands. These are detected via transient binding of fluorescently labeled imager strands. (ii) Representative pseudo-color DNA-PAINT super-resolution image of the large DNA origami platform featuring two ligand attachment sites at 20 nm distance. Ligand (cyan) and platform (red) positions were imaged consecutively by Exchange-PAINT [19]. Scale bar, 50 nm. (iii), Cross-sectional histogram of ligand positions from DNA-PAINT localizations summed up from100 individual DNA origami platforms
3.12 SLB Preparation
Planar supported lipid bilayers (SLB) form spontaneously on a hydrophilic glass surface upon addition of small unilamellar vesicles (SUVs, ∅ = 20–100 nm), which can be easily generated from dried lipid mixtures via bath sonication. We use a lipid mixture containing 98% 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) and 2% 1,2-dioleoyl-sn-glycero-3-[(N-(5-amino-1-carboxypentyl)iminodiacetic acid)succinyl] (nickel salt) (DGS Ni-NTA) to create a mobile SLB that allows for protein attachment via poly-histidine tags.
-
1.
Add 48.6 μL of POPC (10 mg/mL) and 13.8 μL DGS Ni-NTA (1 mg/mL) to a Fiolax® borosilicate test tube to achieve a 49:1 molar ratio and a total of 500 μg of lipid.
-
2.
Fully evaporate the chloroform under a gentle stream of inert gas (e.g., nitrogen) inside a chemical hood until a lipid film has formed.
-
3.
For preparing a 5× stock suspension of lipid vesicles (0.5 mg/mL), add 1 mL of PBS 10× to the dried lipid film and gently resuspend the lipids until the suspension assumes a milky appearance. Firmly seal the test tube (with, e.g., Parafilm) and sonicate the lipid suspension in the test tube for at least 15 min at 20 °C until the lipid suspension has cleared up. Best results are achieved by placing the test tube into the center of a whirl with the surface of the lipid suspension ~3 mm below the water-air interface. The temperature of the water bath should be above the phase transition temperature of the lipid mixture.
-
4.
The 5× lipid vesicle stock suspension can be stored for 2–4 days at 4 °C. Dilute 1:5 with PBS 10× for further use.
-
5.
Ensure glass coverslips are completely dry. Place coverslips into a plasma cleaner, evacuate for at least 10 min to a pressure of 0.2–0.3 mbar before igniting the plasma (pale purple color). Clean for 3 min at a radio frequency power of maximally 30 W. If treatment with an O2 plasma does not yield satisfactory results, immerse coverslips in Piranha solution prior plasma cleaning (1:1 mixture of concentrated sulfuric acid and 30% hydrogen peroxide) for 30 min followed by thorough rinsing with ddH2O).
-
6.
Remove the tape of a sticky-slide 8-well chamber and gently press the cleaned coverslip onto the chamber.
-
7.
Flip the chamber, add 220 μL of the vesicle suspension per well and incubate for 10 min at 24 °C.
-
8.
Carefully rinse the SLB with 20 mL PBS 1× using a 25 mL serological pipette. From a full well, remove 330 μL to leave 350 μL PBS 1× in the well. SLBs can be stored overnight at 4 °C.
3.13 SLB Functionalization
-
1.
Thaw cholesterol-DNA and incubate it on the SLB at a final concentration of 0.1 μM for 60 min at 24 °C.
-
2.
Remove excessive cholesterol-DNA by gently washing the SLB with 10 mL PBS 1× supplemented with 1% BSA. This washing step is crucial for proper attachment of DNA origami to the SLB. Residual cholesterol-DNA in solution will promote formation of DNA origami aggregates.
-
3.
To achieve a surface density of ~1 pMHC per μm2 on the SLB, dilute 5 nM DNA origami solution 1:200 in PBS 1× and add 3 μL to the SLB. Allow to hybridize the elongated staple strands at the bottom side of the DNA origami to complementary cholesterol-DNA in the SLB for 60 min at 24 °C. The surface density of DNA origami structures on the SLB increases nonlinearly with increasing their concentration in solution. The actual density of fluorescently labeled pMHC should always be determined following the protocol given in Subheading 3.16. As a rough guideline, for ~10 pMHC per μm2, dilute 5 nM DNA origami 1:10 in PBS 1× and add 5 μL to the SLB; for ~100 pMHC per μm2, use 20 μL of a 25 nM DNA origami solution.
-
4.
Remove unbound DNA origami by rinsing the SLB with 10 mL PBS 1×.
-
5.
Add His10−tag ICAM-1 and His10-tag B7–1 to a concentration of 5 nM each to the SLB and incubate for 75 min at 24 °C to achieve a surface density of 100 molecules per μm2 for each protein. Rinse with 10 mL PBS 1×. Replace PBS 1× with imaging buffer (HBSS) before cell imaging experiments. Following the precise order of incubation steps on the SLB is critical for achieving efficient and homogeneous decoration of the SLB with all components. Changing the order will decrease DNA origami attachment yields.
3.14 Diffusion Analysis of DNA Origami Structures
-
1.
Set up TIR illumination and focus onto the DNA origami-decorated SLB (see Note 3).
-
2.
Record multiple time-lapsed movies (≥10 movies) at different locations of the SLB for adequate sampling. We typically use an illumination time (till) of 3 ms and a delay of 7 ms between images, yielding tlag = 10 ms based on an expected mobility of DNA origami of <1 μm2s−1. To limit photobleaching, we recommend using laser intensities ≤1 kWcm−2.
-
3.
For analysis, localize single molecules to acquire their positions and connect them to trajectories (e.g., via an open-source software such as ImageJ and the Trackmate plugin). Determine the mean square displacements (MSD) averaged over all trajectories as a function of time lags between images (tlag = till + tdelay). Calculate the diffusion coefficient D from the first two time-lags according to Eq. 1 (Fig. 4a), with σxy being the localization precision. For more details, we refer to [11].
DNA origami characterization. (a) Mobility of DNA origami structures on SLBs. Mean square displacement values extracted from single-molecule tracking experiments are plotted as a function of time lags. Assuming pure Brownian motion, the diffusion coefficient D is derived by considering only the first two data points. (b) Determining the fraction of DNA origami structures devoid of pMHC via two-color co-localization. DNA origami functionalized with AS635P-labeled pMHC are pre-stained with YOYO-1 to label all DNA origami structures. Representative TIRF images of DNA origami on a SLB are shown. Green open circles indicate signals detected in both color channels; red open circles indicate signals detected only in one channel. (c) Determining the number of labeled pMHCs on a DNA origami structure via single molecule brightness analysis. A representative TIRF image of divalent DNA origami platforms bearing two modifications sites for pMHC attachment and the corresponding brightness distributions ρ on SLBs is shown. The detected signals are fitted and the brightness distribution is deconvolved into monomer and multimer contributions (see Subheading 3.16). Scale bar, 2 μm
3.15 Determining the Fraction of DNA Origami Structures Devoid of pMHC
Two-color colocalization of all DNA origami on the SLB (by labeling them with the DNA-intercalating dye YOYO™-1 iodide (YOYO-1)) and fluorescently labeled pMHC (pMHC-AS635P) allows to determine the fraction of SLB-attached DNA origami, which fail to harbor any ligand (Fig. 4b) (see Note 4). For this, the microscope setup should be equipped with two cameras or a beam splitting device (Optosplit II or else) in the emission beam path. Alternatively, two consecutive images acquired with alternating excitation can be recorded on a single camera chip with a delay not longer than 10–20 ms (considering a diffusion coefficient of 0.25 μm2 s−1).
-
1.
Incubate 100 μL (or as needed) of a 5 nM solution of functionalized DNA origami with 100 nM YOYO-1 for 45 min at 24 °C.
-
2.
Follow steps 2–4 from Subheading 3.9, spinning at 4 °C instead of 24 °C. This requires using 7000 g after PuB pre-incubation and 10,000 g for the removal of YOYO-1.
-
3.
Invert the filter, add 50 μL PuB to a fresh clean tube and spin at 5000 g for 4 min at 4 °C. Adjust the final concentration of DNA origami to 5 nM with PuB. Fluorescently stained DNA origami can be stored in Biopur®tubes 1–2 days at 4 °C. Unbound YOYO-1 leads to pronounced background signals on the SLB. If this interferes with the imaging of fluorescently labeled pMHC in the red color channel, we recommend storing the DNA origami solution over night at 4 °C, which will decrease the amount of residual YOYO-1 in solution due to adsorption to the DNA LoBind® tubes.
-
4.
For a surface density of ~1 pMHC per μm2 on the SLB, dilute 5 nM of pMHC-functionalized and YOYO-1-stained DNA origami 1:200 in PBS 1× and add 3 μL to the SLB containing cholesterol-DNA for 60 min at 24 °C.
-
5.
Remove unbound DNA origami by washing with 10 mL PBS 1× supplemented with 1% BSA.
-
6.
Prepare a coverslip featuring immobilized fluorescent multicolor beads (TetraSpeck, Life Sciences) to correct for chromatic aberrations between the two color channels. Record multiple images at different positions using TIR illumination.
-
7.
For the DNA origami sample, record ≥10 two-color images at different positions on the SLB using TIR illumination at laser intensities of 1–3 kWcm−2 and 0.05 kWcm−2 for the red and blue laser, respectively.
For analysis, determine single-molecule positions in both color channels to calculate the relative shift and stretch of the two channels to each other. Count signals in the two channels as colocalized when they occur within a distance of 240 nm of each other, indicated by green open circles in Fig. 4b. The fraction of DNA origami carrying at least one ligand, fbright, can be determined by comparing the number of signals in the red color channel (AS635P-pMHC) that colocalize with signals in the blue color channel (DNA origami, YOYO-1), Ncoloc, to the total number of detected blue signals, Ntotal.
3.16 Determining the Number of pMHCs on a DNA Origami Structure
For DNA origami functionalized at two or more sites, the number of pMHCs per DNA origami can be determined by comparing the brightness of a single DNA origami with the brightness of a single labeled pMHC molecule (Fig. 4c). Note that DNA origami that are devoid of pMHC are invisible in this experiment.
-
1.
Prepare SLBs bearing pMHC-functionalized DNA origami and perform one-color imaging as described in Subheading 3.15 steps 4, 5, and 7.
For analysis, determine the position, integrated brightness B, full width at half-maximum (FWHM) and the local background of individual signals in the images [12, 13]. Routines to perform this analysis are available in MATLAB (see Code availability). Here, brightness values B of functionalized DNA origami decorated with a single ligand are used as a monomer source to calculate the probability density function (pdf) of monomers, ρ1(B).
Based on the independent photon emission process, pdfs of N colocalized emitters can be determined by a series of convolution integrals (Eq. 3):
By applying a weighted linear combination of these pdfs, the brightness distribution of a mixed population of monomers and multimers can be calculated via Eq. 4:
ρ(B) is determined by pooling the brightness values of the detected signals. Individual weights of contributing pdfs can be determined by applying a least-square fit to Eq. 4, assuming \( \sum \limits_{N=1}^{N_{\textrm{max}}}{\alpha}_N=1 \). We recommend a minimum of 800 brightness values for obtaining robust fitting results. Accounting for DNA origami carrying no ligand yields the final fraction of DNA origami carrying N ligands (αN, corrected):
3.17 Determination of pMHC Surface Density
Determine the pMHC surface density on each SLB before using biointerfaces for experiments. Prepare an additional SLB without DNA origami to determine the background signal. In case of multivalent DNA origami, also prepare an SLB bearing monomeric pMHC (e.g., DNA origami functionalized with a single pMHC) to determine the single molecule brightness.
-
1.
Determine the monomer brightness as in Subheading 3.16 steps 1 and 2. Use an SLB featuring pMHCs at a low density (<1 pMHC per μm2).
-
2.
For each biointerface prepared for further experiments as well as the sample used for background correction, record ≥10 bulk images at different locations on the SLB using TIR illumination at laser intensities of 1–3 kWcm−2.
Note: To avoid photobleaching, the exposure time can be reduced for recording bulk images. This must be taken into account in step 4.
-
3.
Analyze the recorded images of biointerfaces with, for example, the open-source software ImageJ. Select a rectangular ROI and calculate the integrated intensity for each sample. Use the same ROI for the background sample to determine the average background intensity for the chosen ROI.
-
4.
To calculate the surface density of pMHC in molecules per μm2, ρpMHC, divide the background-corrected bulk fluorescence average pixel intensity (Itotal – IB) determined in step 2 by the average single-molecule brightness B determined in step 1 and multiply with the number of pixels per μm2 as in Eq. 6.
4 Notes
-
1.
For proper SLB attachment as well as for achieving highest functionalization yields, it is recommended to adhere to the following DNA origami design rules: introduction of reoccurring crossovers every 1.5 turns (=16 bp), corresponding to an inter-helical gap of 1 nm; removal of staple strands along potentially interfering edges to avoid base stacking; 3′-end elongation at sites chosen for ligand as well as cholesterol attachment; insertion of at least two thymidine bases (T) as linkers closest to the DNA origami; and at least 17 complementary bases for all attachment sites to ensure stable DNA hybridization [14,15,16,17]. Not all positions on a DNA origami structure are equally well suited for functionalization, mostly due to varying incorporation efficiencies of staple strands [18]. DNA origami structures with unknown incorporation efficiencies can be characterized via DNA-PAINT, as has been done for the 100 × 70 nm DNA origami platform [18]. We have previously determined and optimized the efficiency of each step of the functionalization process using two-color colocalization analysis (see Subheading 3.15 and Note 4) [10].
-
2.
All modified DNA should be HPLC grade and stored at 50 μM in TE buffer at −20 °C.
For easier handling, we recommend to prepare for each DNA origami layout a separate master mix (MM) consisting of all core staple strands, a surface attachment mix (SAM) consisting of all elongated staple strands at the bottom side, and a ligand attachment mix (LAM) consisting of all elongated staple strands at the top side at a concentration of 50 μM, which can be stored at −20 °C for up to 12 months. The pH of individual components should be verified regularly and set to pH 8.0 to ensure high folding yields. Mix FoB with MgCl2 directly before use only! A tenfold excess of each core staple strand DNA and a 50-fold excess of each elongated/modified DNA over the M13mp18 scaffold concentration is recommended. A larger excess of DNA will not increase the yield of correctly folded and functionalized DNA origami but will complicate the purification process to remove unbound DNA. Whenever handling fluorophores or fluorescently labeled proteins/DNA, protect from light. Use LoBind® tubes for DNA handling and Biopur® tubes for DNA storage.
-
3.
In all single-molecule imaging experiments, care should be taken to avoid bleaching. For optimum imaging conditions, we recommend using fluorophores with superb photostability, high brightness, and negligible unspecific binding/sticking. AlexaFluor™555 is our fluorophore of choice for single molecule experiments; Abberior Star 635P can be used as an alternative if a far-red emitting dye is needed. However, pay attention to nonspecific interaction of this dye with components of the T-cell plasma membrane. To limit photobleaching, we recommend using an aperture in a conjugated plane to the sample plane. The aperture should be set to coincide with the homogeneously illuminated region of interest (ROI), so that fluorescently labeled DNA origami are not pre-bleached before entering the ROI via diffusion.
-
4.
We have tested different DNA intercalating dyes (e.g., PicoGreen™, Sybr™ Gold, Sybr™ Green, YOYO™-3, TOTO™-3) for their suitability for single molecule tracking of DNA origami, but only YOYO-1 offered sufficiently bright signals at low background. Due to its broad emission spectrum, YOYO-1 cannot be used in conjunction with AF555-pMHC; we therefore resort to using AS635P-pMHC for two-color colocalization experiments. Alternatively, a stepwise approach of two consecutive colocalization experiments can be employed: (i) First, we determine the hybridization efficiency of the biotinylated V sequence (Fig. 1a, step 1) using biotin-DNA-AS635P and YOYO-1. (ii) In a next step, we determine the functionalization efficiency with pMHC using biotin-DNA-AS635P and AF555-pMHC (Fig. 1a, step 3). For more details, we refer to [10], where we have determined and optimized the efficiency of each step of the functionalization process using two-color colocalization analysis. In our experience, the overall efficiency of functionalization at a particular modification site with a pMHC molecule is ~70%.
Code Availability
The MATLAB software package for brightness analysis is available via Gitlab: https://github.com/schuetzgroup/TOCCSL_analysis.
References
Fairhead M, Krndija D, Lowe ED, Howarth M (2014) Plug-and-play pairing via defined divalent streptavidins. J Mol Biol 426:199–214. https://doi.org/10.1016/j.jmb.2013.09.016
Howarth M, Liu W, Puthenveetil S, Zheng Y, Marshall LF, Schmidt MM, Wittrup KD, Bawendi MG, Ting AY, Liu W, Puthenveetil S, Zheng Y, Marshall LF, Schmidt MM, Wittrup KD, Bawendi MG, Ting AY (2008) Monovalent, reduced-size quantum dots for imaging receptors on living cells. Nat Methods 5:397–399. https://doi.org/10.1038/nmeth.1206
Huppa JB, Axmann M, Mörtelmaier MA, Lillemeier BF, Newell EW, Brameshuber M, Klein LO, Schütz GJ, Davis MM (2010) TCR-peptide-MHC interactions in situ show accelerated kinetics and increased affinity. Nature 463:963–967. https://doi.org/10.1038/nature08746
Xie J, Huppa JB, Newell EW, Huang J, Ebert PJR, Li Q-JJ, Davis MM (2012) Photocrosslinkable pMHC monomers stain T cells specifically and cause ligand-bound TCRs to be “preferentially” transported to the cSMAC. Nat Immunol 13:674–680. https://doi.org/10.1038/ni.2344
Schnitzbauer J, Strauss MT, Schlichthaerle T, Schueder F, Jungmann R (2017) Super-resolution microscopy with DNA-PAINT. Nat Protoc 12:1198–1228. https://doi.org/10.1038/nprot.2017.024
Rothemund PWK (2006) Folding DNA to create nanoscale shapes and patterns. Nature 440:297–302. https://doi.org/10.1038/nature04586
Douglas SM, Marblestone AH, Teerapittayanon S, Vazquez A, Church GM, Shih WM (2009) Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res 37:5001–5006. https://doi.org/10.1093/nar/gkp436
Hellmeier J, Platzer R, Eklund AS, Schlichthaerle T, Karner A, Motsch V, Schneider MC, Kurz E, Bamieh V, Brameshuber M, Preiner J, Jungmann R, Stockinger H, Schütz GJ, Huppa JB, Sevcsik E (2021) DNA origami demonstrate the unique stimulatory power of single pMHCs as T cell antigens. Proc Natl Acad Sci 118:e2016857118. https://doi.org/10.1073/pnas.2016857118
Shaw A, Benson E, Högberg B (2015) Purification of functionalized DNA origami nanostructures. ACS Nano 9:4968–4975. https://doi.org/10.1021/nn507035g
Hellmeier J, Platzer R, Mühlgrabner V, Schneider MC, Kurz E, Schütz GJ, Huppa JB, Sevcsik E (2021) Strategies for the site-specific decoration of DNA origami nanostructures with functionally intact proteins. ACS Nano 15:15057–15068. https://doi.org/10.1021/acsnano.1c05411
Wieser S, Schütz GJ (2008) Tracking single molecules in the live cell plasma membrane-Do’s and Don’t’s. Methods 46:131–140. https://doi.org/10.1016/j.ymeth.2008.06.010
Moertelmaier M, Brameshuber M, Linimeier M, Schütz GJ, Stockinger H (2005) Thinning out clusters while conserving stoichiometry of labeling. Appl Phys Lett 87:1–3. https://doi.org/10.1063/1.2158031
Schmidt T, Schutz GJ, Gruber HJ, Schindler H (1996) Local stoichiometries determined by counting individual molecules. Anal Chem 68:4397–4401. https://doi.org/10.1021/ac960710g
Seeman NC, Sleiman HF (2018) DNA nanotechnology. Nat Rev Mater 3:17068. https://doi.org/10.1038/natrevmats.2017.68
Wagenbauer KF, Engelhardt FAS, Stahl EK, Hechtl VK, Stömmer P, Seebacher F, Meregalli L, Ketterer P, Gerling T, Dietz H (2017) How we make DNA origami. Chembiochem 18:1873. https://doi.org/10.1002/cbic.201700377
Berger RML, Weck JM, Kempe SM, Liedl T, Rädler JO, Monzel C, Heuer-Jungemann A (2020) Nanoscale organization of FasL on DNA origami as a versatile platform to tune apoptosis signaling in cells:1–20. https://doi.org/10.1101/2020.07.05.187203
Veneziano R, Moyer TJ, Stone MB, Wamhoff EC, Read BJ, Mukherjee S, Shepherd TR, Das J, Schief WR, Irvine DJ, Bathe M (2020) Role of nanoscale antigen organization on B-cell activation probed using DNA origami. Nat Nanotechnol 15:716–723. https://doi.org/10.1038/s41565-020-0719-0
Strauss MT, Schueder F, Haas D, Nickels PC, Jungmann R (2018) Quantifying absolute addressability in DNA origami with molecular resolution. Nat Commun 9:1600. https://doi.org/10.1038/s41467-018-04031-z
Jungmann R, Avendano MS, Woehrstein JB, Dai M, Shih WM, Yin P (2014) Multiplexed 3D cellular super-resolution imaging with DNA-PAINT and exchange-PAINT. Nat Methods 11:313–318. https://doi.org/10.1038/nmeth.2835
Acknowledgments
This work has been supported by research grants of the Austrian Science Fund (FWF project V538-B26 (ES)) and the European Council-sponsored Innovative Medicines Initiative T2EVOLVE (J.H.).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2023 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Hellmeier, J., Platzer, R., Huppa, J.B., Sevcsik, E. (2023). A DNA Origami-Based Biointerface to Interrogate the Spatial Requirements for Sensitized T-Cell Antigen Recognition. In: Baldari, C.T., Dustin, M.L. (eds) The Immune Synapse. Methods in Molecular Biology, vol 2654. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-3135-5_18
Download citation
DOI: https://doi.org/10.1007/978-1-0716-3135-5_18
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-3134-8
Online ISBN: 978-1-0716-3135-5
eBook Packages: Springer Protocols