Abstract
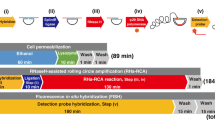
Direct-geneFISH is a Fluorescence In Situ Hybridization (FISH) method that directly links gene presence, and thus potential metabolic capabilities, to cell identity. The method uses rRNA-targeting oligonucleotide probes to identify cells and dsDNA polynucleotide probes carrying multiple molecules of the same fluorochrome to detect genes. In addition, direct-geneFISH allows quantification of the cell fraction carrying the targeted gene and the number of target genes per cell. It can be applied to laboratory cultures, for example, enrichments and phage infections, and to environmental samples. This book chapter describes the main steps of the direct-geneFISH protocol: probe design and synthesis, the “core” direct-geneFISH protocol and lastly, microscopy and data analysis.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Barrero-Canosa J, Moraru C, Zeugner L, Fuchs BM, Amann R (2017) Direct-geneFISH: a simplified protocol for the simultaneous detection and quantification of genes and rRNA in microorganisms. Environ Microbiol 19(1):70–82
Wagner C, Reddy V, Asturias F, Khoshouei M, Johnson JE, Manrique P et al (2017) Isolation and characterization of Metallosphaera turreted icosahedral virus, a founding member of a new family of archaeal viruses. J Virol 91(20):e00925
Moraru C, Lam P, Fuchs BM, Kuypers MMM, Amann R (2010) GeneFISH—an in situ technique for linking gene presence and cell identity in environmental microorganisms. Environ Microbiol 12(11):3057–3073
Allers E, Moraru C, Duhaime MB, Beneze E, Solonenko N, Barrero-Canosa J et al (2013) Single-cell and population level viral infection dynamics revealed by phageFISH, a method to visualize intracellular and free viruses. Environ Microbiol 15(8):2306–2318
Amann R, Fuchs BM (2008) Single-cell identification in microbial communities by improved fluorescence in situ hybridization techniques. Nat Rev Microbiol 6(5):339–348
Greuter D, Loy A, Horn M, Rattei T (2016) ProbeBase—an online resource for rRNA-targeted oligonucleotide probes and primers: new features. Nucleic Acids Res 44(D1):D586–D589
Moraru C, Allers E (2014) Fluorescence in situ hybridization (FISH) for the identification and quantification of microorganisms. In: Skovhus TL, Caffrey SM, Hubert CRJ (eds) Applications of molecular microbiological methods. Caister Academic Press, Wymondham, pp 181–192
Fontenete S, Guimarães N, Wengel J, Azevedo NF (2016) Prediction of melting temperatures in fluorescence in situ hybridization (FISH) procedures using thermodynamic models. Crit Rev Biotechnol 36(3):566–577
Moraru C, Moraru G, Fuchs BM, Amann R (2011) Concepts and software for a rational design of polynucleotide probes. Environ Microbiol Rep 3(1):69–78
McQuin C, Goodman A, Chernyshev V, Kamentsky L, Cimini BA, Karhohs KW et al (2018) CellProfiler 3.0: next-generation image processing for biology. PLoS Biol 16(7):e2005970
Daims H, Lücker S, Wagner M (2006) Daime, a novel image analysis program for microbial ecology and biofilm research. Environ Microbiol 8(2):200–213
Wetmur JG (1991) DNA probes: applications of the principles of nucleic acid hybridization. Crit Rev Biochem Mol Biol 26(3–4):227–259
Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H (1992) Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst Appl Microbiol 15(4):593–600
Pernthaler A, Pernthaler J, Amann R (2002) Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl Environ Microbiol 68(6):3094–3101
Pernthaler A, Pernthaler J, Amann R (2004) Sensitive multi-color fluorescence in situ hybridization for the identification of environmental microorganisms. In: Kowalchuk GA, de Bruijn FJ, Head IM, Akkermans AD, van Elsas JD (eds) Molecular microbial ecology manual. Springer, Dordrecht, pp 2613–2627
Pavlekovic M, Schmid MC, Schmider-Poignee N, Spring S, Pilhofer M, Gaul T et al (2009) Optimization of three FISH procedures for in situ detection of anaerobic ammonium oxidizing bacteria in biological wastewater treatment. J Microbiol Methods 78(2):119–126
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Barrero-Canosa, J., Moraru, C. (2021). Linking Microbes to Their Genes at Single Cell Level with Direct-geneFISH. In: Azevedo, N.F., Almeida, C. (eds) Fluorescence In-Situ Hybridization (FISH) for Microbial Cells. Methods in Molecular Biology, vol 2246. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-1115-9_12
Download citation
DOI: https://doi.org/10.1007/978-1-0716-1115-9_12
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-1114-2
Online ISBN: 978-1-0716-1115-9
eBook Packages: Springer Protocols