Abstract
This chapter describes the usage of the program ARGweaver, which estimates the ancestral recombination graph for as many as about 100 genome sequences. The ancestral recombination graph is a detailed description of the coalescence and recombination events that define the relationships among the sampled sequences. This rich description is useful for a wide variety of population genetic analyses. We describe the preparation of data and major considerations for running ARGweaver, as well as the interpretation of results. We then demonstrate an analysis using the DARC (Duffy) gene as an example, and show how ARGweaver can be used to detect signatures of natural selection and Neandertal introgression, as well as to estimate the dates of mutation events. This chapter provides sufficient detail to get a new user up and running with this complex but powerful analysis tool.
You have full access to this open access chapter, Download protocol PDF
Similar content being viewed by others
Key words
1 Overview
The ancestral recombination graph (ARG) can be considered the holy grail of statistical population genetics. The ARG represents the history of a collection of related genome sequences, in terms of the coalescence events by which segments of genomes trace to common ancestral segments and the historical recombination events that cause patterns of ancestry to differ from one genomic site to the next (see Chapter 1 for more introduction to these concepts). Provided the sequences under study are orthologous and co-linear—meaning that they trace to a common ancestral sequence without genomic duplications or rearrangements—the ARG is a complete description of their evolutionary relationships. Moreover, in statistical terms, the ARG provides a highly compact and precise description of the correlation structure of such a collection of sequences. Importantly, the ARG naturally defines a set of recombination breakpoints, a set of haplotypes, and a genealogy for each non-recombining interval in the genome—all objects that are useful starting points for countless population genetic analyses.
Many questions in applied population genetics can be reframed as questions about ARG structure. For example:
-
Recombination rate estimation. Recombination rates can be estimated by simply counting recombination events and dividing by the total branch-length of the ARG.
-
Estimation of allele ages or mutation rates. Mutation events can easily be mapped to branches within the ARG by maximum parsimony, enabling straightforward estimation of allele ages and mutation rates.
-
Local ancestry inference. The local ancestry structure of an admixed individual (i.e., which genomic segments derive from which distinct source populations) can be determined by tracing the individual’s two diploid lineages in the ARG and identifying the source population with which each genomic segment clusters, as well as the recombination events that terminate these segments.
-
Demography inference. More general information about demographic history (such as population sizes, migration rates, and divergence times) is also embedded in the ARG. A demographic model can fairly easily be estimated from a known ARG by making use of the counts of coalescence events within and between populations.
-
Detection of sequences under selection. Natural selection can be detected by identifying local distortions in the ARG, for example, unusual clusters of coalescence events or extremely deep times to most recent common ancestry.
In practice, however, the true ARG is impossible to know with certainty. The “ARG space,” consisting of every possible ancestral history of a set of genomes, is astronomically large, and the information in genome sequences is insufficient to choose a specific ARG above all others. But, given a model of coalescence, recombination, and nucleotide substitution, it is possible to compute the probability of an observed data set under particular ARGs, and it will generally be true that some ARGs are much more likely to have produced the data than others. The approach taken by ARGweaver is to sample from the posterior distribution of ARGs, given a collection of genome sequence data and a reasonable set of modeling assumptions. This approach is computationally expensive, and it has the drawback of producing a complex and unwieldy output—a collection of potential ARGs, none of which is exactly correct, but which, in the aggregate, reflect certain properties of the true ARG. Nevertheless, as we will show, this approach can be extremely powerful, potentially providing insights into the structure of the data and the evolutionary history of the sample that are not easily obtained using simpler methods. In this chapter we will discuss how ARGweaver works, how and when a user might want to apply it, and what can be done with sampled ARGs once they have been obtained.
1.1 What Is an ARG?
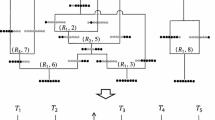
An ARG represents all ancestral relationships among a collection of genomes (see Fig. 1). If n is the number of (haploid) genomes under study (usually from \( \frac{n}{2} \) diploid individuals), then at the present day, there are n lineages in the ARG. As we trace these lineages back in time at a particular genomic location, we will find that distinct lineages gradually coalesce into shared ancestral lineages, until all n lineages have found a single most recent common ancestor. These coalescence events define a tree known as a genealogy that fully describes the evolutionary relationships among the present-day genomes at the locus in question.
(a) Schematic of an ARG with four lineages in the present, and two ancestral recombination events along a region of length L. Tracing the lineages upwards from present day, two lineages merge when a coalescence event is encountered, whereas a lineage splits into two when a recombination event is encountered at a particular breakpoint (b2 or b3). The ARG continues tracing the history backwards until all lineages have reached a common ancestor. (b) An alternative view of the ARG depicted in A, showing the local tree between each pair of recombination breakpoints. The dotted lines on the tree show the recombination event which transforms the tree on the left side of the breakpoint into the tree on the right side. (c) The data underlying this ARG, where only derived alleles at variant sites are shown. Figure adopted from [23]
However, recombination events in the history of the sample can cause the genealogy to change from one genomic location to the next. Looking backward in time, a recombination at a particular genomic location has the effect of splitting a lineage into two, with one path representing the evolutionary history to one side of the breakpoint and another path representing the history to the other side. The ARG captures these recombination events together with the coalescence events. As one follows a lineage upward in the ARG, that lineage may either merge with another lineage, representing a coalescence event, or it may split into two lineages, representing a recombination event (Fig. 1a). In the case of recombination events, the junction in the ARG is also labeled with the genomic position of the recombination (this information is not relevant for coalescence events).
Based on these labels for recombination events, one can extract a local tree for any position in the genome from the ARG. First, one identifies the lineage associated with each present-day sample. These lineages are traced backward through the ARG, and coalescences between them are noted. When a recombination event is identified, one of the two possible paths is selected based on the relationship of the position in question to the annotated recombination breakpoint. Specifically, if the position is to the left of the breakpoint, then the left path is taken; and if the position is to the right of the breakpoint, then the right path is taken. (Because recombination breakpoints by definition occur between nucleotides, one of these two cases must hold.) Thus, the paths from the present-day samples to the root will coalesce only, never splitting, and therefore must define a tree. Furthermore, the tree will be the same for all genomic positions between two recombination breakpoints, differing only between positions on opposite sites of a breakpoint.
Another way to think about the ARG is that it defines a series of operations on trees along the length of a chromosome. As one walks along a chromosome from left to right, the local tree remains fixed until a recombination breakpoint is encountered, and then that tree is altered to form a new tree, in the specific manner defined by the change in path at the corresponding recombination node in the ARG (Fig. 1b). The ARG, therefore, can be thought of as being interchangeable with a sequence of local trees and the associated recombination events that transform each tree to the next. In practice, this is the representation of the ARG assumed by the Sequentially Markov Coalescent (SMC′) and used by ARGweaver, and in this chapter we will generally treat the ARG as a collection of trees and recombination events. Nevertheless, it should be noted that this representation does not strictly capture all of the information in the ARG. The full ARG also describes “trapped genetic material” that falls between two linked ancestral loci, but is not passed on to any present-day sample. Ignoring this trapped material substantially simplifies modeling and inference algorithms, with what appear to be only minor costs in accuracy [12, 14, 23].
1.2 Why Would You Want to Estimate an ARG?
As discussed above, if the ARG could be estimated accurately and easily, it would be useful for almost every question in population genetics. In practice, of course, there are limitations in the accuracy of inferred ARGs, and they require substantial time and effort to obtain. So, when does it make sense to take the trouble to run ARGweaver, instead of making use of simpler or more standard population genetic summary statistics and tools? Some reasons to consider sampling ARGs with ARGweaver include:
-
Trees/genealogies. ARGweaver estimates explicit genealogies (with branch lengths) along the genome, considering both patterns of local mutation and local linkage disequilibrium. It may be particularly interesting to inspect trees at particular regions suspected to be under selection or to have experienced introgression.
-
Times/dates. These trees allow the timings of various events to be estimated, including times to most recent common ancestry, other coalescence times, and the ages of derived alleles. If desired, posterior expected values of these times can be computed by averaging over the sampled trees.
-
Ancient introgression. ARGweaver is a powerful method for detecting introgression and identifying specific introgressed haplotypes, particularly ancient introgression events that conventional methods may miss (e.g., [6]).
-
Bayesian treatment of uncertainty. Unlike many simpler methods, ARGweaver attempts to fully account for the uncertainty in the ARG given the sequence data and an evolutionary model, by sampling from a posterior distribution of ARGs. This approach can mitigate biases from the inference method in addressing biological questions of interest.
-
Flexibility in addressing “custom” evolutionary questions. By producing explicit ARGs, ARGweaver allows almost any evolutionary question to be addressed, including unusual ones not easily addressed with standard summary statistics (For example: at what fraction of sites do individuals A and B coalesce with one another before either coalesces with individual C? What is the average TMRCA for genes of functional category X? Are recombination events more likely to occur in introns or intergenic regions?)
-
Technical limitations of the data. ARGweaver can accommodate unphased data, low-coverage sequences, archaic samples, and other unusual data types that may not be easy to analyze using other methods.
1.3 Practical Considerations
ARGweaver is designed to run on genome sequencing data for small to moderate numbers of individuals—anywhere from 2 to a maximum of about 100. These individuals should be unrelated but come from the same species or from recently diverged species (such as humans and chimpanzees). Phasing of diploid genome sequences is not necessary—ARGweaver can phase “on the fly,” integrating over possible phasings—but the algorithm converges faster and, in some cases, performs better on phased data (depending on the rate of phasing errors). Similarly, ARGweaver can be used on low-coverage sequencing data, making use of genotype probabilities to weight the observed bases, but high-coverage sequence data is always preferable.
In gauging the feasibility of ARG inference, it is important to recognize that the processes of mutation and recombination are opposing forces in reconstructing an ARG. The more mutations there are, the more information there is to guide the inference of tree topologies (genealogies). Recombination events, however, break up the sequences into smaller blocks, effectively limiting the information for tree inference in each block. Thus, the quality of ARG inference depends on the ratio of mutation to recombination rates per nucleotide position. In human data, this ratio is close to one, but recombination events tend to be concentrated in recombination hotspots, which makes the effective ratio greater than one for most of the genome. ARGweaver appears to work quite well in this setting. Nevertheless, the method works better when this ratio is even higher, and it will break down if this ratio falls significantly below one. Another consideration is ARGweaver’s assumption of at most one recombination event per site (see below), which generally appears to have little effect but could lead to biased estimates in cases of particularly high recombination rates, large sample sizes, large evolutionary distances, or large effective population sizes. Finally, because ARGweaver depends on haplotype-scale information for inference, it is generally not useful for short sequences, deriving, for example, from RAD-seq or a de novo short-read assembly.
In terms of the number of genomes analyzed, the “sweet spot” for ARGweaver is generally between a handful of individuals and a few dozen. As the number of genomes increases, more approximate models (such as the Li and Stevens model [8]) or conventional population genetic summary statistics become increasingly accurate and informative, and the relative advantage of using ARGweaver over other methods decreases. In addition, the run time and size of the ARGweaver output increase with the number of genomes, and these factors become prohibitive with more than about 100 samples. Running ARGweaver genome-wide generally requires breaking the genome into chunks of a few megabases and running ARGweaver in parallel on each chunk using a computer cluster. When running ARGweaver genome-wide is not a realistic possibility, it may still be of interest to apply ARGweaver to specific genomic regions of interest, such as candidate selective sweeps or introgressed regions. It may also be useful to run ARGweaver on subsets of the available genome sequences, for example, to shed light on genealogy structure, ancient introgression, or allele age—features ARGweaver may estimate more accurately than other methods.
Another practical consideration is that while ARGweaver’s output is richly informative, it is not straightforward to interpret. The program does come with tools to compute various local summary statistics from sampled ARGs, including times to the most recent common ancestor, allele ages, and distances between samples. But many less standard analyses will require custom programs to extract the desired information from ARGs or local genealogies.
1.4 ARGweaver Algorithm Overview
ARGweaver uses a Markov chain Monte Carlo (MCMC) algorithm to sample ARGs at frequencies proportional to their probability, conditional on the observed DNA sequence data (X) and the model parameters (θ). The MCMC algorithm starts with an initial ARG, G0, and then repeatedly removes a subset of the ARG and resamples that subset from an appropriate conditional probability distribution. This process generates a sequence of ARGs, G0, G1, …, Gm, where m is the total number of iterations of the algorithm. Although G0 may be a poor guess with low probability, by sampling each new Gi according to the appropriate distribution, the chain will eventually converge to the desired distribution—i.e., for sufficiently large i, Gi will represent a draw from the posterior distribution over ARGs given the data and the model, P(Gi|X, θ). In practice, it is customary to plot the posterior probability as a function of the iteration number, i, observe the point at which it ceases to trend upward and becomes stable, and then to discard the ARGs sampled before this point (from what is known as the “burn-in” of the MCMC algorithm).
Even once the algorithm has converged, successive samples Gi and Gi+1—while they both represent samples from the posterior distribution—are not independent samples. Rather they are strongly correlated, since only part of the ARG is resampled on each step of the algorithm. Therefore, in order to achieve a distribution of nearly independent ARGs—both to save space and processing time, and to better assess the variance of estimates derived from the samples—it is useful to “thin” the chain, recording only every jth sample (the default thinning parameter in ARGweaver is j = 10). After discarding the initial “burn-in” and performing thinning, the ARGs Gi that remain can be stored and treated as a collection of samples representative of the distribution of ARGs given the data and the model, P(G|X, θ).
The technical details of the ARGweaver algorithm will not be reviewed here (see ref. 23), but the main idea is to remove a single haploid genome from the ARG, and then to “thread” this genome back through the ARG, by sampling both its coalescence points with the remaining sequences and the associated recombination points. There is also another, slightly more complicated, version of this threading operation, called “subtree threading,” that resamples internal branches in genealogies, and is essential for ARGweaver to efficiently explore the full space of possible ARGs. In both cases, a hidden Markov model (HMM) is used to efficiently sample new coalescent points for the new lineage across the chromosome. This HMM depends on several key modeling assumptions, which are important for users to understand, and which, therefore, will be reviewed in the next section.
1.4.1 ARGweaver Model and Assumptions
The HMM underlying ARGweaver depends on the following assumptions:
-
SMC′or SMC: The Sequentially Markov Coalescent model [14] or the closely related SMC′ [12] is assumed. These models posit that the distribution over genealogies at each nucleotide position directly depends only on the genealogy at the previous position, not on the genealogies at positions further upstream—a feature known in probability theory as the Markov property, after the Russian mathematician Andrey Markov. More formally, the SMC and SMC′ assume that the genealogy at position i + 1 is independent of the genealogies at positions 1, …, i − 1, given the genealogy at position i. The SMC′ slightly improves on the original SMC (see ref. 12 for details). The differences between these models are not important here, and the choice of model seems to have only a subtle effect on the inferred ARGs. While the SMC′ is technically more accurate, the SMC model may be considerably faster on data sets with large numbers of samples. ARGweaver therefore allows the user to choose either model (SMC by default, --smc-prime for the SMC′).
-
Discrete time: All recombination and coalescent events are assumed to occur at a predefined collection of discrete time points. The total number of time points, K, can be chosen by the user (using --ntimes <K>) and can be arbitrarily large, with the ARGweaver model approaching a continuous-time model as K approaches infinity. However, the computational complexity of the threading algorithm is proportional to K2, so, in practice, K must be kept modest in size. The default value of K in ARGweaver is 20. The time points are uniformly spaced on a logarithmic scale, so that they are more closely clustered at recent time points, when there are more lineages and coalescence rates are larger. The algorithm forces all lineages to coalesce by the final time point, tK.
-
No more than one recombination event between neighboring nucleotides. For simplicity, the algorithm permits at most one recombination event at every “step” along the sequence, meaning between two adjacent nucleotide positions. This assumption means that adjacent genomic positions must either have identical genealogies or ones that differ by a single recombination event. In practice, this assumption is minimally restrictive, because the information about genealogies comes primarily from variable sites, which tend to be sparse along the genome. If ARGweaver should need to account for multiple recombination events between variable sites, it typically can spread those events across a series of intervening invariant sites with minimal impact on accuracy. If the data are such that multiple recombinations between neighboring sites occur frequently, then it is likely that the haplotype structure is too broken down to make use of ARGweaver.
-
Population size known: ARGweaver assumes that the effective population size Ne (which determines the coalescence rate) is provided by the user. In the simplest case, a single global value of Ne can be provided. But ARGweaver can accommodate different values of Ne for different discrete time intervals. Values of Ne can typically be obtained from the literature or estimated from the same data using one of the many available programs for inferring demographic histories (such as SMC++ [29], PSMC [7], MSMC [27, see also Chapter 7], G-PhoCS [3], and diCal [28]). Note the user-provided values of Ne define a “prior” for coalescence rates in ARGweaver, so it is not necessary for them to be perfectly estimated; ARGweaver will consider the data together with this prior distribution in sampling coalescence events.
-
Mutation and recombination rates known. The ARGweaver model also depends on pre-defined mutation and recombination rates. These rates can be assumed to be constant across the genome, or variable rates can be provided in a position-specific map along the genome. These values are also “priors” in the same sense as the population size (see above).
-
Jukes-Cantor model of base substitution. ARGweaver makes use of a Jukes-Cantor model for nucleotide substitutions. This model assumes that all nucleotide substitutions are equally probable—an obvious oversimplification, but one that seems to have minimal costs at the close evolutionary distances typically considered by ARGweaver. The symmetries inherent in the Jukes-Cantor model can be exploited to optimize the likelihood calculations in ARGweaver.
2 Ancient Hominins Analysis
In the remainder of this chapter we will set up, and then walk through, an analysis of real sequence data using ARGweaver. We will use three high-quality ancient hominin genome sequences that are freely available: the Altai Neandertal [21], Vindija Neandertal [22], and Denisovan [15] genome sequences, as well as a diverse set of 14 human genomes that were sequenced to high coverage for the Altai Neandertal paper [21]. All steps of the analysis will be described in detail, with code snippets, and the complete set of commands required to replicate the analysis is provided in the companion material for this book.
The data set used in our example is ideal for several reasons. First, the Neandertal and Denisovan genome sequences provide an exciting opportunity to examine many interesting aspects of human history and adaptation. Neandertals and Denisovans are sister groups of archaic hominins that diverged from humans roughly 600 kya, and then split from each other around 400 kya [22]. Importantly, these divergence times are recent enough that modern and archaic humans share many polymorphic sites across their genomes. As we will show, this shared variation can be informative about evolutionary history. In addition, the genetic evidence strongly suggests multiple cases of interbreeding among these three groups following their initial divergence [15, 21, 24], an intriguing topic that can be examined using ARGweaver. On a more practical level, these genomes have all been sequenced to high coverage and all sequencing reads have been processed consistently. The genotypes are published in standard VCF format with genotype quality and sequencing depth information given at every genomic position with aligned reads, which, as we will discuss, simplifies the set up for ARGweaver.
Before launching into the actual analysis, which is presented in Subheading 5, we will discuss some important preliminaries relating to program installation and file formats (remainder of Subheading 2), model parameters (Subheading 3), and commonly used program options (Subheading 4).
2.1 Pre-requisites
ARGweaver is designed to run under either the Linux or Mac OSX operating system. Windows(c) users may run ARGweaver via a Linux virtual machine. The specific commands for performing the example analysis in this chapter are available as a bash script, provided in the online supplement for this book. Besides a bash shell, the script requires:
-
Python: (http://python.org), used by several ARGweaver scripts.
-
SAMtools [9]: (http://htslib.org), used here for the tabix and bgzip tools, which are useful for indexing and fast retrieval of VCF and BED files.
-
bedops [18]: (http://bedops.readthedocs.io), a useful tool for computing intersections of genomic intervals.
-
PHAST [20]: (http://compgen.cshl.edu/phast), used for computing neutral substitution rates.
-
R (https://www.r-project.org), used for plotting results.
-
The R package “ape” [19] (https://bioconductor.org), used for plotting trees.
-
git (https://git-scm.com) for downloading ARGweaver.
-
g++ (https://gcc.gnu.org) or any C++ compiler for compiling ARGweaver.
2.2 Obtaining and Installing ARGweaver
The first step in our example is to download and install ARGweaver. The program is available at http://github.com/CshlSiepelLab/ARGweaver.git. It can be downloaded and compiled on Linux or Mac machines with the following commands:
git clone https://github.com/CshlSiepelLab/ARGweaver.git cd ARGweaver.git make
These commands create several executables in the bin/ directory, the most important of which is called arg-sample. All the executables are meant to be run from the command line in a Unix shell such as Bash.
Within the ARGweaver software is also suite of R tools useful for plotting ARGweaver results. This package is optional, but was used to create many of the plots in this chapter. It can be installed from the same directory with the command:
R CMD INSTALL R/argweaver
2.3 Sequence File Format
The main data required by ARGweaver is sequence data for every individual. ARGweaver accepts Variant Call Format (VCF) files [2], provided that they are indexed (this can be done with the command tabix -p vcf file.vcf.gz, which creates a file file.vcf.gz.tbi). A single VCF file containing all samples may be provided with the argument --vcf; if only a subset of the individuals are to be used, they can be specified with the --subsites option. Or, if the genotypes are in multiple VCF files, a list of these files can be given with the option --vcf-files. In our example, there is a single VCF file for each individual, so we use the second option.
It is worth noting that VCF input has some limitations when used with ARGweaver. Namely, ARGweaver will ignore any phasing information in the VCF, and sites with insertion/deletion polymorphisms or more than two alleles. If the data is phased, the SITES format will have to be used (see below).
2.4 SITES Format
ARGweaver has its own sequence data format, called SITES format, which is used both as an alternative input format, as well as an output format for sampling phase. All lines in SITES files are tab-delimited. The first line starts with the string “NAMES” and then lists the name of every haploid genome (two per individual). The second line starts with the string “REGION” and is followed by the chromosome name, start position, and end position. All subsequent lines contain two columns: the position of a variant site and a string giving the observed alleles at this site in each of the genomes, in the order given on the first line. Importantly, any position not listed in the file is considered invariant across samples. Here is a short example of a sites file with the two Neandertal individuals and two variant sites on chromosome 2:
NAMES Altai_1 Altai_2 Vindija_1 Vindija_2 REGION 2 24000001 24010000 24000417 GGCG 24008883 TTTA
2.4.1 Phasing Options
For an ARG to be fully defined for diploid organisms, the genotypes at heterozygous positions must be “phased” into two distinct haploid genome sequences. The low-cost, short-read sequencing methods most widely in use, however, generally produce unphased data, in which the chromosomal origin of each allele at a heterozygous site is unknown. Thus, an important consideration in running ARGweaver is how to address phasing.
ARGweaver can either accept predefined haplotype phases or it can treat the phase as unknown and sample possible phasings as it samples ARGs. The program’s default behavior is to assume haploid genome sequences are fully specified (phased input). The option --unphased causes ARGweaver to sample the phase instead. In unphased mode, whenever ARGweaver re-threads a leaf branch, it does so by integrating over possible phasings of the individual corresponding to that leaf. After the threading is complete, it resamples the phase for this individual conditional on the threading choice. This new phase is retained until the next time a leaf for the same individual is re-threaded. The phase sampling step is fast and does not contribute significantly to the run time of each sampling iteration, but it may delay overall convergence of the algorithm.
As noted above, the program does not read phase information from VCF files, so the --unphased option is implicit when using VCF input. In this case, the program internally creates two haploid lineages, <ind>_1 and <ind>_2, for each diploid individual, <ind>, listed in the VCF file. (Throughout this chapter, we assume diploid input to ARGweaver; in principle, ARGweaver could be used with haploid input, but the assumed model of recombination is best matched to sexually reproducing species.) These haploid labels will be used in some of the output files of ARGweaver. With SITES input and the --unphased option, the program requires the user to indicate the haploid pairs corresponding to each individual. They will be detected automatically if the genomes are named with the convention <ind>_1 and <ind>_2. Otherwise, the option --unphased-file <file.txt> may be used, where <file.txt> has two columns with haploid sample names, and each row corresponds to a single individual.
In unphased mode, ARGweaver will output the current phased data (in SITES.gz format) every time an ARG is sampled. This explicit phasing information makes it possible to map mutations onto branches of the sampled ARGs, among other features. It has the additional benefit of allowing ARGweaver to function as an ARG-based computational phasing method. In practice, however, other existing phasing methods are more efficient, and are likely to achieve greater accuracy, especially if they are able to leverage large reference panels of phased genomes [10].
Nevertheless, even when the data has been pre-phased by another computational method, it may still be worthwhile to use the --unphased option. The reason is that the error rates from computational phasing methods can be quite high, and in unphased mode ARGweaver may be able to “correct” phasing choices that are incompatible with the ARGs it samples. In this case, the pre-phased data can still be passed to ARGweaver in SITES format and will be used for initialization, so convergence will be much faster than with unphased input.
2.5 Masked Regions
Whether using VCF or SITES input, ARGweaver assumes that any site which does not appear in the input file is invariant. An “invariant” site in ARGweaver is one in which all individuals have been sequenced and are confidently called homozygous for the same allele (usually the reference allele). This absence of genetic variation is informative about the ARG. For example, a genomic region with many invariant sites will tend to have short branches in the inferred ARG, because fewer mutations are expected on shorter branches.
On the other hand, some sites have unknown genotypes, for example, due to low sequencing depth, poor sequence quality, or poor alignability. For ARGweaver, an unknown genotype means something quite different from an invariant site—it suggests missing data, meaning that no information is provided about the ARG at that genomic location. At a region with many unknown sites, the sampled trees will tend to reflect the neutral coalescent model provided to ARGweaver, because there will be little data available to override this “prior.”
Therefore, it is essential to distinguish between “invariant” and “unknown” sites in the input files to ARGweaver. Making this distinction often requires some additional effort, because other population genetic methods often do not depend strongly on it, and many data sets are not processed in a way that tracks this difference. In particular, genotypes that are unknown should be “masked,” either by using the genotype NN or by using masking options (described below), so that ARGweaver knows to integrate over all possible genotypes at those positions. Unmasked sites that are not specified in the input can then reasonably be assumed to be invariant.
ARGweaver supports several kinds of masking. For regions with poor alignability (such as repeat regions), it is customary to mask out the entire region across all individuals. This can be done by providing ARGweaver with a BED-formatted file indicating the regions to mask, and using the option --maskmap <mask_file.bed>. (Note that unlike VCF and SITES files, BED files have zero-based start coordinates.) On the other hand, some regions have poor genotype quality only in particular individuals, for example, due to low sequencing coverage or read quality. In these cases, the regions can be delineated in an auxiliary file which is specified using the option --ind-maskmap <ind_mask_file.txt>. This file should have two columns, giving the name of each individual and the name of a file containing a BED-formatted mask specific to that individual. Additional options include --mask-cluster <a,b>, which will mask any region of length b that has ≥ a variant sites (possibly indicating alignment errors or mutational hotspots); --vcf-min-qual <Q>, which will mask any genotype with quality less than Q (for VCF files with quality scores); or --vcf-genotype-filter <filter>, which can mask genotypes based on any keys used in the VCF genotype field. For example, --vcf-genotype-filter ~DP<10;DP>50;GQ<10~ will mask sites where the depth is less than 10 or greater than 50, or where the genotype quality is less than 10.
In our example analysis, we use a union of several mappability and uniqueness filters developed for the ENCODE project [30]. Details for where these filters were obtained can be found in the online resource. We also use --vcf-min-qual 30 --mask-cluster 2,5. These filters seem sufficient for our illustration, however other filters may be needed for a thorough, careful analysis. In particular, it may be important to mask CpG sites, which have unusually high mutation rates.
2.5.1 Genomic vs Variant VCFs
Many population genomic data sets are now available in VCF format, and it may be tempting to run ARGweaver directly on such a data set. However, VCF is a common file type used for a wide variety of purposes, and it is critical to understand what criteria were used for inclusion or non-inclusion of sites in the files before analyzing them. In particular, VCF files often contain only those positions where high-confidence variants are detected. As discussed above, this convention will make it impossible to distinguish unknown and invariant sites. Thus, more preparation will be necessary for a proper analysis with ARGweaver.
Further processing of such incomplete VCF files generally requires returning to the alignments from which the VCF files were derived. If those alignments are available in the form of a BAM file, then the necessary information can be extracted in a fairly straightforward manner. If they are not available, it may be necessary to regenerate them from the raw reads. Once a BAM file is in hand, the best option is usually to re-run a genotype caller (such as GATK [31]) on the BAM file to generate more complete VCF files including most likely genotypes at every site, as well as quality scores, sequencing depth, and genotype probabilities. A possible shortcut, adequate for many purposes, is to use bamtools [1] to extract the sequencing depth per site from the BAM file, and use the depth as a proxy for genotype quality. For example, if a position is not in the VCF file but has a sequencing depth greater than some cutoff (perhaps 20), then it is very likely invariant. In practice, this thresholding can be accomplished by first creating a BED file for each individual containing the regions with sequencing depth below the desired cutoff, and then using the --ind-maskmap option to mask these regions.
In our example, we are lucky to be using VCF files that provide genotype probabilities and confidence scores at every location having aligned reads. However, care must still be taken to deal with these files correctly. ARGweaver assumes that any site absent from a VCF file is invariant, but in this case those sites are actually unknown. To correct this assumption, we create a BED file containing all the regions absent from the VCF for each individual, and use the --ind-maskmap option to specify that these regions should be masked in their respective genomes. This is done with the bedops tool [18], and the commands are shown in the example script that comes with this chapter.
2.5.2 Genotype Probabilities
If a genome has only been sequenced at low coverage, there may be too many errors in the genotypes to produce meaningful ARGs. Nevertheless, it is possible to run ARGweaver on low-quality data by having it weight possible genotypes by their probabilities of being correct. There are two ways to specify these probabilities. First, if VCF files are annotated with PL (phred-likelihood) or GL (genotype-likelihood) fields, then the option --use-genotype-probs may be used to integrate over the possible genotypes. Second, genotype probabilities can be encoded directly into the SITES file. In this case, each row (following the header) may have an additional 4n columns, in the order \( {p}_{1,A},{p}_{1,C},{p}_{1,G},{p}_{1,T},{p}_{2_A},\dots, {p}_{n,T} \), where pi,b is the probability of the ith haploid genome having base b. If these columns are present, then --use-genotype-probs is implied.
The use of genotype probabilities will slow down ARGweaver, and therefore this feature should only be used if absolutely necessary. There is a modest computational cost, of course, in taking the genotype probabilities into account. The larger issue, however, is that the use of genotype probabilities causes almost every site to be considered “variant,” which prohibits the use of site compression (see Subheading 4.3). Nevertheless, genotype probabilities may be useful for low-coverage data when the scale of the analysis is not too large.
3 Choosing Model Parameters
ARGweaver assumes fixed rates of mutation, recombination, and coalescence (based on population sizes), which must be specified by the user. As noted above, these parameters can be thought of as defining a “prior” distribution for ARGs, which can be overcome by consideration of the data in determining the “posterior” samples produced by the program. However, these prior estimates can have an appreciable influence on the sampled ARGs, so they should be set as accurately as possible.
3.1 Mutation Rates
ARGweaver can accept a single mutation rate to be used across the entire genomic region that is being analyzed (with the option --mutrate <rate>), or it can use a specified map of mutation rates (--mutmap <ratefile.bed>). If using a rate map, the file specifying the map should have four columns: chromosome, start coordinate (0-based), end coordinate, and the rate. The rates should be specified in units of expected mutations per base pair per generation.
The mutation rate is particularly important for calibrating the timing of ancestral events. If the given mutation rate is off by a factor of m, then the estimated ages of events will tend to be off by a factor of 1∕m, so that too high a mutation rate will make events seem to have happened much more recently than they actually did. After a period of controversy [26], estimates of mutation rates for humans have stabilized in recent years, but there is still considerable debate about the best average rates to use for evolutionary analyses [16, 25]. In addition, mutation rates are known to vary across species and along the genome in each species, which further complicates their use in ARGweaver.
For our example analysis, we address these issues by using levels of divergence between several closely related primates (human, chimpanzee, gorilla, orangutan, and gibbon) to estimate relative mutation rates in sliding 100 kb windows. We first mask out conserved regions of the genome in order to estimate the neutral substitution rate, which should be proportional to the average mutation rate. Then, we scale all the relative rates so that the average rate is 1.45e − 8 mutations per base pair per generation [17]. The online resource for this chapter contains the full script for obtaining these rates, as well as the rates themselves.
3.2 Recombination Rates
Recombination rates are specified in ARGweaver in units of the probability of a recombination between two neighboring bases per generation. As with mutation rates, the rate may be assumed constant across the region (--recombrate <rate>), or a map of rates may be provided to the program (--recombmap <ratefile.bed>).
As discussed earlier, there is an important interplay between the mutation and recombination processes in the ARGweaver model. ARGweaver always tries to find ARGs that best fit the data given the model. If too high a recombination rate is used, then ARGweaver may overfit the data, so that it samples as many recombination events as needed to produce trees that allow for minimal numbers of mutations at polymorphic sites. Conversely, an unrealistically low rate will lead to ARGs containing too few recombination events, resulting in local trees that are incompatible with the site patterns in the data (this will be seen as a large number of “noncompats” in the output stats file). Thus, it is important to specify the recombination rates as accurately as possible.
In humans, there are many estimates of recombination rates; for our example analysis we will use one based on a collection of African-American genomes [5]. For other species that may not have existing maps, a genome-wide estimate from the closest model organism should be sufficient.
It is worth noting that, for reasons of computational efficiency, the mutation and recombination map should generally only be specified at modest levels of resolution along the genome sequence. The reason is that ARGweaver must recompute the transition and emission probabilities of its HMM at all positions at which the mutation or recombination rates change, as well as at those at which the local tree changes. Thus, if the rates change more frequently than the local trees, there will be a considerable increase in the run time of the algorithm. For this reason, in our example analysis we smooth out the recombination rates by taking the average across 5 kb windows. The mutation rates were calculated in larger windows so they are already smooth.
3.3 Population Size
The prior model for ARGweaver assumes that all samples are drawn from a single, panmictic population. The panmixia aspect of the prior is weak in the sense that actual structure in the population is usually evident in the data and will be reflected in the sampled ARGs. At the same time, the population size does determine the prior rate at which branches coalesce, so an incorrect prior may skew certain features of the sampled ARGs, such as the relative coalescence times between samples, and the relative rates of candidate recombination events.
The simplest way to specify population size is as a constant-sized population. A quick estimate for the population size can be obtained using Watterson’s estimator. If S is the number of segregating sites over L nucleotides in n haploid genomes, and μ is the mutation rate per base pair per generation, then Watterson’s estimator for the diploid effective population size is:
A slightly better method, for the purposes of ARGweaver, is to use the nucleotide diversity (otherwise known as pi), which is computed as the average number of pairwise differences between any two haploid genomes per base-pair. If using VCF input, this calculation can be accomplished using VCFTools [2] with the command vcftools --site-pi. The nucleotide diversity can be divided by 4μ to obtain an estimate of the diploid effective population size, N.
For most populations, a more realistic model allows for a changing population size over time. Programs such as SMC++ [29], MSMC [27, see also Chapter 7], PSMC [7], G-PhoCS [3], or diCal [28]) can be used to obtain estimates of a demographic history that includes such changes. The option --popsize-file <popsize_file.txt> can then be used to specify the corresponding history in ARGweaver. The specified popsize file should have two columns: a time (in generations) and a diploid population size. The times should be increasing, with the first time being zero. An example is shown below:
0 10000 1500 200 2500 20000
This file would represent a bottleneck scenario where there is a population of size 10,000 for the past 1500 generations, but between generations 1500 and 2500 ago, the population size was only 200. Before 2500 generations ago, the population size was 20,000. Note that ARGweaver will round all times in the file to the nearest discrete time point used by the model.
Our example analysis illustrates some of the inherent shortcomings in assuming a single population. The sequence data we will analyze consists of humans sampled from across the globe, including non-Africans (whose populations endured a severe bottleneck associated with the out-of-Africa event, followed by a rapid recent expansion), Africans (whose effective population size is larger, and more stable over time), and ancient hominins (whose effective population sizes are much smaller than those of humans). There simply is no single history which would be a good fit for our data set. In this case, we will simply forge ahead with ARGweaver’s default population size of 10,000, which is about the correct order of magnitude for most of our lineages over most of their shared history. However, it is critical that we keep our model misspecification in mind as we interpret our results. In a real analysis, we would probably eventually want to use simulations to understand the implications of our over-simplifying assumptions. At the same time, the fact that we do estimate ARGs that appear to be reasonable in most respects demonstrates that ARGweaver is somewhat robust to the choice of population size.
3.4 Time Discretization
Before running ARGweaver, it is worth thinking about the time discretization scheme and adjusting it to ensure it is appropriate for the analysis at hand. There are three options to consider. First, the number of time points can be changed with the option --ntimes <ntime>, with the default number of points being 20. While more resolution may be desired, the running time will increase proportionally to the square of this number.
Second, the option --maxtime <time> indicates the maximum time point in the model, in units of generations. All lineages are forced to coalesce by this time, so it usually makes sense to choose a very ancient time. The default in ARGweaver of 200,000 generations is about 20 times the effective human population size and a reasonable choice for most human analyses.
The third relevant option is --delta <delta>. The times are distributed on a scale so that recent time points are more close together than distant points. This convention allows for greater resolution on recent time scales, when there are usually more coalescence events (since there are more distinct lineages). The distribution is controlled by the delta (δ) parameter, where which bring the points closer together at recent times when it is set to larger values. The exact formula for setting the time points is: \( t(i)=\left(\exp \left(\frac{i}{K-1}\log \left(1+\delta {t}_{max}\right)\right)-1\right)/\delta \), for K time points and i ∈{0, 1, …, K − 1}. Very small values of δ(< 1∕tmax) will yield roughly linear distribution of times, whereas very large values δ will place the first few time points so close together that they represent fractions of a generation. The default value of δ = 0.01 produces a reasonable distribution for the default tmax of 200,000 generations. The discrete times are written to the terminal and log file at the start of an ARGweaver run, and it is advisable to inspect these values and possibly adjust δ (by restarting the run) as necessary.
It is important to carefully consider your goals when deciding how to set maxtime and delta. If you are interested in recent history, then you may want to increase delta, and your choice of maxtime may not be crucial. If you are interested in balancing selection or deep coalescences, it will be important to choose a larger maxtime and possibly a smaller delta as well.
4 Other Options
4.1 Sampling Frequency
Another decision to be made is how frequently to sample from the MCMC chain. The option --sample-step <n> tells ARGweaver to output the ARG sampled on every nth step of the MCMC algorithm. If any of the individuals are unphased, then the program will also output the corresponding phased samples.
As mentioned above, it is customary to “thin” MCMC samples to reduce autocorrelation between the final samples, but it is difficult to know how much thinning will be required prior to an ARGweaver run. Some have argued that thinning is inefficient and unnecessary, and that most properties of the distribution can be better estimated using the full sample [11]. In the case of ARGweaver, however, there is a substantial cost associated with storing and processing each sampled ARG, and adjacent ARGs in the chain are very highly correlated, so some degree of thinning is justified. We will use the default sampling frequency of 10, and return to the issue of autocorrelation as we interpret the results.
4.2 Ancient Samples
If the samples are not all from present day (as with the Neandertals and Denisovan in our example), their ages (in generations before the present) can be specified to ARGweaver. The computed ARGs will then have shorter branches for these samples than for the modern samples. The ages can be specified in a file with two columns—sample name and age in generations—and passed to ARGweaver with the option --age-file <age_file.txt>. The ages will be rounded to the nearest time point in the model. Any sample not found in the file is assumed to have age zero, and at least one sample must have age zero (so sample ages should be given relative to the youngest sample).
In our example, we use ages of 4206 generations for the Altai Neandertal, 2482 generations for the Denisovan sample, and 1793 generations for the Vindija Neandertal, corresponding to ages of 122 kya, 72 kya, and 52 kya, respectively [22], and an assumed generation time of 29 years.
4.3 Site Compression
Site compression is usually important for keeping ARGweaver’s run time manageable. The option --compress-seq <c> will compress blocks of c sites together, resulting in a speed-up of the code of approximately a factor of c. The breakpoints between each block are chosen in a flexible manner so that there is no more than one variant site in the same block. If a block contains a variant site, then the new “compressed site” takes on the site pattern of its variant site; otherwise the compressed site is invariant. The mutation and recombination rates are also increased by a factor of c, since they reflect per-site rates. (This rate inflation is done internally by arg-sample; the user should provide rates per uncompressed base pair.)
There are several issues to consider with this option. Compression will fail (with a program abort soon after stating) if variant sites are too close together to compress at the requested level. Compression also causes a loss of resolution; if --compress-seq 50 is used, then the breakpoints in the ARGs will occur at most every 50 base pairs. Most importantly, recall that ARGweaver assumes that only one recombination event can occur between any two sites. This assumption applies to compressed sites also, so its effect is magnified as the compression factor increases. Therefore, if compression is too high, ARGweaver may not be able to place enough recombination events between variant sites, resulting in poor ARG estimates.
One way to think about how much compression is allowable is to consider the distribution of distances between variant sites, ignoring singleton sites, which contain no topological information. In our data set, non-singleton variant (NSV) sites occur, on average, about every 300 bases. Therefore, one might be tempted to select a compression factor of 50, which would allow an average of ∼6 recombinations between each pair of NSV sites. However, bear in mind that the distance between NSV sites is approximately exponentially distributed, which means that at an average distance is 300, only ∼15% of NSV sites are more than 50 base pairs apart. We therefore will use a compression factor of 10 in our example, which should allow about 97% of pairs of NSV sites to have more than one recombination between them.
In general, we recommend using compression conservatively, so that multiple recombinations are still possible between most pairs of adjacent NSVs. Too high compression will lead to a high number of “noncompats” in the .stats file (described in the next section), so the compression factor may need to be adjusted if this is observed.
5 Running ARGweaver
With all preliminaries addressed, we are now ready to run ARGweaver and work through our example analysis. The command to run ARGweaver is as follows:
arg-sample --vcf-files vcf_files.txt \ --region $region \ --vcf-min-qual 30 \ --subsites inds.txt \ --maskmap filter.bed.gz \ --mask-cluster 2,5 \ --ind-maskmap ind_mask_files.txt \ --age-file sample_ages.txt \ --mutmap subst_rate_autosome.bed.gz \ --recombmap recomb_rate_autosome.bed.gz \ --compress-seq 10 \ -o $outdir/out
As ARGweaver runs, it produces several output files, all with names having the prefix specified by the -o option. These files include:
-
A log file (<outroot>.log) , which records the same output that is written to the terminal, including details about the model and data, progress, time, and memory usage, etc., as the iterations continue.
-
A stats file (<outroot>.stats), consisting of one row per MCMC iteration, with columns including:
-
prior: log probability of the sampled ARG given the model
-
likelihood: log probability of the data given the sampled ARG
-
joint: total log probability of the ARG and the data (prior + likelihood)
-
recombs: number of recombination events in the sampled ARG
-
arglen: total length of all branches summed across sites
-
noncompats: the number of variant sites that cannot be explained by a single mutation under the sampled ARG
-
-
Sampled ARGs (<outroot>.<iter>.smc.gz) are written at the sampling frequency requested by the option --sample-step. These are in ARGweaver’s SMC format, which is text-readable and lists non-recombining genomic intervals, the tree in each interval (using Newick format), and the recombination events that occur between intervals. The recombination events are described as subtree pruning and regrafting (SPR) events, which define where a particular branch breaks and recoalesces back onto the tree.
-
Phased sites files (<outroot>.<iter>.sites.gz) If the data is unphased, then it will also print the current phased sequence data (in SITES format) for each ARG that is printed. These phased SITES files do not contain information about positions that are masked in all individuals; those regions are written to a file named <outroot>.masked_regions.bed.
5.1 Time/Memory Requirements
ARGweaver requires substantial computation time, but the memory usage is low. In our example, a 1 Mbase region took 6 h to complete 1000 MCMC iterations, with a maximum memory usage of 135 Mbytes. The program does not support multithreading. Rather, parallelization is usually achieved by running many genomic segments at the same time on different CPU cores.
5.2 Monitoring Convergence
The stats file produced by ARGweaver can be used to monitor ARGweaver’s convergence. Figure 2 shows an example of how statistics change as the sampler reaches equilibrium (known as “stationarity” in the MCMC literature). In this example, the relevant statistics seem to become fairly stable after about 600 iterations, so we will use 600 for the “burn-in” for this run.
Traces of various statistics over MCMC iterations. The values should stabilize as the chain reaches convergence; earlier iterations should be discarded as “burn-in.” Values related to the probability of the data, including the prior, likelihood, and joint probability, should increase as the MCMC converges from a poor initial guess to higher probability ARGs. The number of “noncompats” tends to decrease until stabilization, since the true ARG usually explains observed site patterns without requiring multiple mutations. The number of recombinations and length of the ARG may increase or decrease before convergence, depending on the data and the model. In this example, a burn-in of 600 iterations seems adequate
It is also useful to examine the autocorrelation of the statistics in the stats file (after removing the burn-in samples). The autocorrelation of the “joint” statistic is shown in Fig. 3, and it appears that in this case autocorrelation reaches insignificant levels in roughly 20 iterations. Autocorrelations for the other statistics look similar and are insignificant between 20–30 iterations (not shown). Therefore, the sampled ARGs should be thinned to at least every 20th MCMC iteration in order to achieve a sample of effectively independent ARGs. It is not necessary to perform thinning in order to estimate the mean or quantiles of ARG statistics, so we will not do that here. Still, it is useful to keep the thinning interval in mind, in order to compute how many effectively independent samples we have. If we run 1000 MCMC iterations, and discard the first 600 as burn-in, and use a thinning interval of 20, we are left with an effective sample size of only 20. This size may be sufficient for inspecting example trees, but is not enough to obtain good estimates of derived quantities such as coalescence times. So, in this case we may want to continue running the MCMC chain for (at least) an additional 1600 iterations in order to end up with 100 effectively independent samples.
The autocorrelation in the “joint” likelihood statistic for the run shown in Fig. 2, as plotted by the R function “acf.” The region between the blue lines represents the 95% confidence interval for no correlation
5.2.1 Resuming a Run
In our original command, we ran the MCMC for the default number of iterations, which is 1000. It is easy to resume a run, by adding the option --resume and specifying the final number of iterations desired, for example, --iters 2600. In this case it will start from iteration 1000 and perform 1600 additional iterations.
6 Interpreting Results
When enough iterations have been completed, the fun begins! It is time to look at the ARGs and see what might be learned from them. There are countless ways that one might parse and examine a set of ARGs, and custom code may eventually be required for a specific analysis. However, there are some tools in ARGweaver that can be used to get started.
Many of the plots we will show in this and following sections were created by the R package that comes with ARGweaver. We will not show the code to create the plots in this chapter, but it is in the companion material that accompanies this book. The R package Gviz [4] was used to create the gene annotation plots.
6.1 Leaf Trace Plots
Leaf trace plots were introduced in the ARGweaver paper [23] as way to visualize the ARG. In these diagrams, the leaves of the local trees, as they change along the genome sequence, are drawn as horizontal lines, with the vertical distance between neighboring lines proportional to the distance between adjacent leaves in the local tree (see top panel of Fig. 4). Leaf traces should be interpreted with caution, because there are many possible leaf trace plots for the same ARG (depending on arbitrary choices made in ordering the leaves), and the distance between non-adjacent lines in the leaf trace plot is not directly interpretable. Furthermore, the leaf trace is drawn for a single ARG, rather than showing the distribution across sampled ARGs. Nevertheless, the leaf trace is an intuitive graphical description of an ARG that can be used to survey its overall structure.
Leaf trace plots and key statistics for the region surrounding the DARC gene. The highlighted region shows the location of SNPs that define different haplotypes, some of which provide resistance to malaria. The recombination rate is shown in units of log(recombs/bp/generation) and is computed across all individuals; the solid gray line shows the prior recombination rates, and the black line shows posterior expected values. The other statistics are shown for different population subsets. TMRCA gives the time to the most recent common ancestor; RTH is the “relative TMRCA half-time,” which describes the minimum time for half the lineages to coalesce, normalized by the TMRCA. Blue lines plot African human genomes; red lines plot non-African humans, and black lines in the leaf trace (top panel) represent the ancient hominins. Shading around all lines represent 90% confidence intervals
A leaf trace plot can be created by first running the arg-layout executable on a single SMC file. Then, the “plotLeafTrace” function in the ARGweaver R package can plot the resulting file.
Leaf traces around the DARC gene are shown in the top panel of Fig. 4. The first feature that is apparent is that the plot changes quickly along the x-axis in some regions, and more slowly in others, reflecting the posterior estimate of the local recombination rate. The vertical height of the plot also gives a quick indication of the total height of the local trees along the region. That is, a large “spread” of the traces indicates a deep (ancient) time to most recent common ancestry (TMRCA), whereas a small spread indicates a shallow TMRCA. If the traces are colored by population of origin, the leaf trace can also provide an idea of the level of population structure in the data. The leaf trace in our example suggests that the DARC gene is in the middle of a low-diversity region with a relatively high recombination rate.
6.2 Computing Basic ARG Statistics
After examining leaf trace plots, the next way to explore the ARG is to look at various statistics across the genomic region. The first step is to convert the ARGs into a more convenient format. We currently have an SMC file for every sampled ARG, named <outprefix>.0.smc.gz, …, <outprefix>.<num_iter>.smc.gz. We use the command smc2bed-all <outprefix>, which will combine all the information into a single sorted and indexed BED file, with columns: chromosome, start coordinate (0-based), end coordinate, MCMC iteration number, and Newick tree (representing the local tree for the ARG sampled in this region and iteration).
Now, the executable arg-summarize can be used to extract statistics. Some of the more useful options to arg-summarize include:
-
--tmrca: time to the most recent ancestor
-
--pi: average distance between any two leaves
-
--branchlen: total tree length
-
--popsize: estimate of diploid population size based on coalescence rates in the local tree
-
--tmrca-half: time at which half the samples find a current ancestor
-
--rth: Relative TMRCA Half life (RTH), defined as the ratio of tmrca-half to tmrca. Unusually low values of this statistic suggest a recent “clustering” of coalescent events, possibly indicating a partial selective sweep [23]
-
--node-dist <leaf1,leaf2>: Distance between leaf1 and leaf2 on the tree
-
--node-dist-all: Like node-dist, but for all pairs of leaves
-
--min-coal-time <ind1,ind2>: Return minimum coalescence time between two individuals (over all four haploid pair combinations)
-
--subset-inds <ind_list.txt>: Before computing statistics, prune all individuals not listed in given file
-
--mean: Rather than reporting statistics for every MCMC iteration, report the mean across iterations for all non-recombining intervals. This applies to all statistics requested (i.e., --tmrca,--pi, etc.)
-
--quantile <q1,q2,...>: Same as --mean, but instead report one or more quantiles of statistics across samples
For example, the command:
arg-summarize -a <outprefix>.bed.gz --tmrca \ --subset africans_inds.txt \ --quantile 0.05,0.5,0.95
would compute the 5%, 50% (median), and 95% quantiles of the time to the most recent common ancestor, in the subset of the ARG only containing individuals listed in the file african_inds.txt.
ARGweaver supports many additional options not described here. A full list can be obtained using the command arg-summarize --help.
Figure 4 shows a plot of several of these statistics (recombination rate, pi, popsize, RTH, TMRCA) in the region surrounding the DARC gene. The highlighted region is known to harbor variants which have reached near-fixation throughout Africa and are thought to provide resistance to malaria; however, this region tends not to be detected by most tests for selective sweeps [13]. Looking at Fig. 4, there are some suggestions of possible positive selection in Africa, such as low estimates for pi, population size, and RTH in the highlighted region. It is possible that the relatively high recombination rate in this region has led to a fast breakdown of haplotype structure, which would make a selective sweep difficult to detect.
It is important when looking at these plots to remember the underlying population structure; a low RTH seems to be fairly common in non-African populations due to the population bottleneck, but is more rare in African populations. Similarly, we expect the local population size estimates for the African genomes to be higher than for the non-Africans. Overall, while the RTH in Africa at the highlighted region is low, it is doubtful that this region would be a significant outlier in a genome-wide scan.
6.2.1 Examining Local Trees
Often one of the most useful ways to gain insight into the ARG is to look directly at the estimated local trees. ARGweaver’s R package comes with some tools to visualize the trees (the package internally makes use the “ape” package [19]). For example, Fig. 5 shows one of the sampled trees at the position of rs2814778, the SNP which defines the African haplotype.
This tree is interesting in several ways. First, it suggests a tight grouping among the African and non-African populations. While this is not particularly unusual for non-Africans (due to the out-of-Africa bottleneck), it is quite rare to observe this level of clustering among Africans. Importantly, the San individual does not cluster with the rest of the Africans, providing a hint as to why the African TMRCA was not unusually low in Fig. 4. In fact, it is known that the FY*O mutation shown in Fig. 5 is not common in the San population, whereas it has reached near-fixation throughout most of the rest of Africa [13].
These observations suggest that perhaps the statistics shown in Fig. 4 do not fully capture the interesting aspects of this tree. Figure 6 shows the same statistics, but also calculated using a subset of Africans that excludes San. This subset (shown in dark blue) is much more of a regional outlier, with unusually low pi, population size, and TMRCA. Notably, the RTH statistic is no longer low in this case; this is because RTH is designed to detect partial sweeps, and the putative sweep is complete in the subset excluding San.
One must of course be cautious about the biases that might result from inspecting the sampled trees, deciding how they are interesting, and then revising the statistical tests to detect these same interesting features. Nevertheless, it is sometimes only by visual inspection and exploration of the sampled trees that the patterns in the data start to become clear. The basic statistics that can be computed with arg-summarize are often too crude to give a clear understanding of the ARG features. For example, TMRCA is unaffected by any feature besides the final coalescence time. The pi statistic is also very heavily dominated by long branches, especially due to the logarithmic timescale used by ARGweaver. Finally, RTH is sometimes informative of partial sweeps; however, it fails to detect complete sweeps or partial sweeps that have not yet hit 50% frequency, and it also will give many false positives if the underlying population is expanding or has experienced a bottleneck.
It is therefore sometimes necessary to “browse” through the trees to get an idea of what interesting signals may exist. In our example, we revised our tests both because of the structure of the observed trees, and because this structure was concordant with the known geographical distribution of the DARC haplotypes. In order to determine if this locus is truly special, it would be necessary to compare its trees/statistics to ones from across the genome, or generated from neutral simulations based on a more realistic demographic model.
6.2.2 Allele Age
The sampled ARGs allow mutations to be mapped to the branches of local trees, which in turn allows the times at which those mutations occurred to be estimated. The arg-summarize program supports computing such “allele ages,” based on a single sites file. However, this program is not designed to perform allele age computations when the data is unphased, as it would need to use the sampled phase corresponding with each sampled ARG to properly map the mutations. This problem can be addressed using a script called allele_age, which will call arg-summarize for each pair of sampled ARG and phase, and report the age for each MCMC iteration.
There are some caveats to interpreting the allele age. First of all, it is typically the case that, at a small fraction of sites, the local tree will not be able to explain a particular variant site with a single mutation (indicating a violation of the “infinite sites” model), so that the mutation time is poorly defined. It is also possible that the assignment of derived/ancestral alleles will not be consistent across all the ARGs; some ARGs may explain an observed variant as a young, low-frequency allele, whereas other samples may flip the ancestral and derived alleles and describe the same variant as an old, high-frequency allele. Also, when a mutation is mapped to a branch, it is equally likely to have arisen anywhere along the branch, so mutations mapped to longer branches will have much more uncertainty in their time estimates than those mapped to short branches.
The allele_age function outputs a number of extra columns to help clarify these issues. For each MCMC sample, it outputs the identity of the inferred derived and ancestral alleles, as well as a flag indicating whether the mutation can be explained by the infinite sites model under the sampled tree. It also outputs both the mean allele age (the midpoint of the branch where it was mapped) and the minimum allele age (the most recent point on the branch).
In our DARC example, we find that on average 4.5% of sites do not obey the infinite sites model. The infinite sites violations tend to be concentrated in the same sites across all MCMC replicates (with 2.6% of sites requiring multiple mutations in > 95% of replicates, and 92% of sites requiring multiple mutations in < 5% of replicates). This rate of infinite-site violations is not unexpected, and is likely due to a number of factors, including low levels of genotyping error accumulating over 17 samples, true instances of multiple mutations (especially at sites with high mutation rates such as CpG sites), model misspecification, or uncertainty in phasing or the ARG.
Looking at the allele underlying the tree in Fig. 5, we estimate that the allele is between 100 and 300 ky old. This large amount of uncertainty is not surprising, as the mutation is mapped to a fairly long branch above the African subtree.
6.2.3 Neandertal Introgression
Careful inspection of the local trees in our example region reveals another interesting feature. Several of the trees downstream from DARC, such as the one shown in Fig. 7, exhibit an atypical placement of one of the Han haplotypes (Han_2), which is clustered tightly with the Neandertal genomes. In other respects, this tree is quite typical. It is therefore possible that this is a region of Neandertal introgression into the Han individual included in our data set.
To investigate further, we can use arg-summarize to compute the minimum coalescence time between the Han individual and each of the ancient hominins (for every sampled ARG, at every genomic location), and look at the distribution of these times. The result is plotted in Fig. 8. We see that there is a region between roughly 159.20Mb and 159.21Mb where the Han genome coalesces with both the Neandertal and the Altai genomes significantly more recently than 500 kya, roughly the minimum to be expected in the absence of introgression. (The threshold of 500 kya generously allows for uncertainty in dating coalescence events; the actual population divergence time is closer to 600 kya.) No other individual besides Han has coalescence times significantly below 500 kya in this region (not shown), indicating that this observation is not likely to be an artifact of using an incorrect local mutation rate. Thus, it seems likely that this is a short introgressed region in the Han.
Minimum coalescence times for the Han individual to each of the three ancient individuals (red: Neandertal, blue: Denisovan; minimum taken across all four haplotype combinations Han_1, Han_2, ancient_1, ancient_2). Times are in ky; shaded colors show 95% confidence intervals across MCMC samples, solid lines give the median. The horizontal line at t=500 ky represents an approximate minimum expected time of coalescence in the absence of introgression, given the divergence time between humans and Neandertal/Denisovans. The highlighted region is putatively introgressed from Neandertal into the Han genome
Another way to examine this region is to look at the site patterns within it. The subsites program can retrieve sites for particular individuals or genomic intervals; we can use the sampled phased SITES file from the ARGweaver run to examine the site patterns. The R package also has a function (plotSites()) to visualize site patterns. Figure 9 shows the site patterns in this putatively introgressed region. It confirms that there are four sites shared by one of the Han haplotypes and at least one Neandertal, which are not shared by any other modern human, making the region a very good candidate for introgression. Most other software for detecting introgression would not confidently find such a short region.
7 Discussion
We hope that this chapter is sufficient to help new users decide whether ARGweaver is an appropriate tool to apply to their data, and to get started with an initial analysis. In our example, we explored a few relatively simple statistics that suggested instances of positive selection and Neandertal introgression near the DARC locus. In other work, we have also shown that ARGweaver can be used to detect balancing and negative selection [23], as well as more subtle patterns of introgression [6]. However, there are many more evolutionary questions that ARGweaver could potentially shed light upon; in fact, one of the most exciting aspects of ARGweaver is that its possible uses have yet to be fully explored. One could imagine delving deeper into the study of natural selection, for example estimating selection coefficients, or distinguishing selection on new variants from selection on standing variation. Beyond natural selection, the ARG could also be useful to detect patterns of population structure, explore genotype/phenotype correlation, phase haplotypes, estimate recombination rates, etc. As we discussed in the introduction, almost all population genetics questions can be framed as questions about ARG structure. In some cases it may be sufficient to compute summary statistics from the ARG using arg-summarize, but in others, it will be necessary to write custom code for analyzing the ARG.
There is no doubt that an ARGweaver analysis requires more time and effort than an analysis based on a typical “off-the-shelf” tool for population genetics. For this reason, ARGweaver generally should not be the first tool that one reaches for when analyzing a new data set. Nevertheless, as we have shown, this additional effort can prove worthwhile for certain kinds of analyses. One such instance is when the genomic data are rare and precious, as is the case with the Neandertal, Denisovan, and other ancient genomes, which is why we highlight the use of those genomes in this chapter. Another circumstance in which ARGweaver may be especially useful is when simpler methods fall short, for example, by being underpowered, or inappropriate in some way for the data or the question at hand. Because ARGweaver utilizes all the genome data, without reducing it to summary statistics, and models the full recombination and coalescence process (within the limitations of the SMC′), it should generally have more statistical power than other methods. In addition, its ability to work on low-quality and/or unphased genomes, and produce full evolutionary histories, makes it a uniquely flexible approach that may fill in gaps left behind by more traditional methods. For these reasons, ARGweaver is a “power tool” many population geneticists may wish to add to their toolboxes.
Change history
27 February 2021
Chapter 2, “Processing and Analyzing Multiple Genomes Alignments with MafFilter,” was previously published without including the Electronic Supplementary Material. This has now been included in the revised version of this book.
References
Barnett DW, Garrison EK, Quinlan AR, Strömberg MP, Marth GT (2011) BamTools: a C++ API and toolkit for analyzing and managing BAM files. Bioinformatics 27(12):1691–1692. https://doi.org/10.1093/bioinformatics/btr174
Danecek P, Auton A, Abecasis G, et al (2011) The variant call format and VCFtools. Bioinformatics 27(15):2156–2158. https://doi.org/10.1093/bioinformatics/btr330
Gronau I, Hubisz MJ, Gulko B, Danko CG, Siepel A (2011) Bayesian inference of ancient human demography from individual genome sequences. Nat Genet 43(10):1031–1034
Hahne F, Ivanek R (2016) Visualizing genomic data using Gviz and bioconductor. Statistical genomics: methods and protocols. Springer, New York, pp 335–351
Hinch AG, Tandon A, Patterson N, et al (2011) The landscape of recombination in African Americans. Nature 476(7359):170–175
Kuhlwilm M, Gronau I, Hubisz MJ, et al (2016) Ancient gene flow from early modern humans into Eastern Neanderthals. Nature 530(7591):429–433
Li H, Durbin R (2011) Inference of human population history from individual whole-genome sequences. Nature 475:493–496
Li N, Stephens M (2003) Modeling linkage disequilibrium and identifying recombination hotspots using single-nucleotide polymorphism data. Genetics 165:2213–2233
Li H, Handsaker B, Wysoker A, et al (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079
Loh PR, Palamara PF, Price AL (2016) Fast and accurate long-range phasing in a UK Biobank cohort. Nat Genet 48:811–816
MacEachern SN, Berliner LM (1994) Subsampling the Gibbs sampler. Am Stat 48(3):188–190
Marjoram P, Wall JD (2006) Fast “coalescent” simulation. BMC Genet 7:16
McManus KF, Taravella AM, Henn BM, et al (2017) Population genetic analysis of the DARC locus (Duffy) reveals adaptation from standing variation associated with malaria resistance in humans. PLOS Genet 13(3):1–27. https://doi.org/10.1371/journal.pgen.1006560
McVean GA, Cardin NJ (2005) Approximating the coalescent with recombination. Philos Trans R Soc Lond B Biol Sci 360:1387–1393
Meyer M, Kircher M, Gansauge MT, et al (2012) A high-coverage genome sequence from an archaic Denisovan individual. Science 338(6104):222–226. https://doi.org/10.1126/science.1224344
Moorjani P, Gao Z, Przeworski M (2016) Human germline mutation and the erratic evolutionary clock. PLoS Biol 14(10):e2000744
Narasimhan VM, Rahbari R, Scally A, et al (2017) Estimating the human mutation rate from autozygous segments reveals population differences in human mutational processes. Nat Commun 8(1):303. https://doi.org/10.1038/s41467-017-00323-y
Neph S, Kuehn MS, Reynolds AP, et al (2012) BEDOPS: high-performance genomic feature operations. Bioinformatics 28(14):1919–1920. https://doi.org/10.1093/bioinformatics/bts277
Paradis E, Claude J, Strimmer K (2004) APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20:289–290
Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A (2010) Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res 20:110–121
Prüfer K, Racimo F, Patterson N, et al (2014) The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505(7481):43–49
Prüfer K, de Filippo C, Grote S, et al (2017) A high-coverage Neandertal genome from Vindija Cave in Croatia. Science 358(6363):655–658. https://doi.org/10.1126/science.aao1887
Rasmussen MD, Hubisz MJ, Gronau I, Siepel A (2014) Genome-wide inference of ancestral recombination graphs. PLoS Genet 10(5):e1004342
Sankararaman S, Mallick S, Dannemann M, et al (2014) The genomic landscape of Neanderthal ancestry in present-day humans. Nature 507:354–357
Scally A (2016) The mutation rate in human evolution and demographic inference. Curr Opin Genet Dev 41:36–43
Scally A, Durbin R (2012) Revising the human mutation rate: implications for understanding human evolution. Nat Rev Genet 13(10):745–753
Schiffels S, Durbin R (2014) Inferring human population size and separation history from multiple genome sequences. Nat Genet 46(8):919–925
Sheehan S, Harris K, Song YS (2013) Estimating variable effective population sizes from multiple genomes: a sequentially Markov conditional sampling distribution approach. Genetics 194(3):647–662
Terhorst J, Kamm JA, Song YS (2016) Robust and scalable inference of population history from hundreds of unphased whole genomes. Nat Genet 49:303–309
The ENCODE Project Consortium (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489:57–74
Van der Auwera GA, Carneiro MO, Hartl C, et al (2013) From FastQ data to high-confidence variant calls: the genome analysis toolkit best practices pipeline. Curr Protoc Bioinformatics 11(1110):11.10.1–11.10.33. https://doi.org/10.1002/0471250953.bi1110s43
Acknowledgements
The ARGweaver software was initially written by Matt Rasmussen, and later substantially extended by Melissa Hubisz. We thank Aaron Stern, Amy Williams, and Ritika Ramani for helpful comments and suggestions. This material is based upon work supported by the National Science Foundation Graduate Research Fellowship to Melissa Hubisz under Grant No. DGE-1650441. Adam Siepel was supported in part by grant GM102192 from the National Institutes of Health.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
1 Electronic supplementary material
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2020 The Author(s)
About this protocol
Cite this protocol
Hubisz, M., Siepel, A. (2020). Inference of Ancestral Recombination Graphs Using ARGweaver. In: Dutheil, J.Y. (eds) Statistical Population Genomics. Methods in Molecular Biology, vol 2090. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-0199-0_10
Download citation
DOI: https://doi.org/10.1007/978-1-0716-0199-0_10
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-0198-3
Online ISBN: 978-1-0716-0199-0
eBook Packages: Springer Protocols