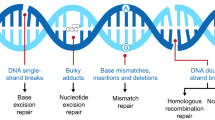
Abstract
Purpose
Deleterious germline BRCA1/2 mutations are among the most highly pathogenic variants in hereditary breast and ovarian cancer syndrome. Recently, genes implicated in homologous recombination repair (HRR) pathways have been investigated extensively. Defective HRR genes may indicate potential clinical benefits from PARP (poly ADP ribose polymerase) inhibitors beyond BRCA1/2 mutations.
Methods
We evaluated the prevalence of BRCA1/2 mutations as well as alterations in HRR genes with targeted sequencing. A total of 648 consecutive breast cancer samples were assayed, and HRR genes were evaluated for prevalence in breast cancer tissues.
Results
Among 648 breast cancers, there were 17 truncating and 2 missense mutations in BRCA1 and 45 truncating and 1 missense mutation in BRCA2, impacting 3% and 5% of the study population (collectively altered in 6%) with cooccurrence of BRCA1/2 in 7 breast cancers. On the other hand, HRR genes were altered in 122 (19%) breast cancers, while TBB (Talazoparib Beyond BRCA) trial-interrogated genes (excluding BRCA1/2) were mutated in 107 (17%) patients. Beyond BRCA1/2, the most prevalent HRR mutant genes came from ARID1A (7%), PALB2 (7%), and PTEN (6%). Collectively, 164 (25%) of the 648 Taiwanese breast cancer samples harbored at least one mutation among HRR genes.
Conclusions
The prevalence of BRCA1/2 mutations was far below one tenth, while the prevalence of HRR mutations was much higher and approached one-fourth among Taiwanese breast cancers. Further opportunities to take advantage of defective HRR genes for breast cancer treatment should be sought for the realization of precision medicine.







Similar content being viewed by others
References
Most common cancer diagnosis globally: breast surpasses lung - medscape Dec 17, 2020. Robson M, Im SA, Senkus E, Xu B, Domchek SM, Masuda N, Delaloge S, Li W, Tung N, Armstrong A, Wu W, Goessl C, Runswick S, Conte P. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N Engl J Med. 2017;377:523-33. URL: https://www.medscape.com/viewarticle/942808. Accessed Mar 17, 2021.
Health Promotion Administration, Ministry of Health and Welfare, Taiwan. Cancer registry annual report, 2017, Taiwan. Taipei: Health Promotion Administration, Ministry of Health and Welfare, Taiwan, 2019.
Department of Statistics, Ministry of Health and Welfare, Taiwan. Cause of death statistics, 2019, Taiwan. https://www.mohw.gov.tw/cp-16-54482-1.html (2019, Accessed 24 Sept 2020).
Kuo CN, Liao YM, Kuo LN, Tsai HJ, Chang WC, Yen Y. Cancers in Taiwan: practical insight from epidemiology, treatments, biomarkers, and cost. J Formos Med Assoc. 2020;119(12):1731–41.
US Preventive Services Task Force, Owens DK, Davidson KW, Krist AH, Barry MJ, Cabana M, Caughey AB, Doubeni CA, Epling JW Jr, Kubik M, Landefeld CS, Mangione CM, Pbert L, Silverstein M, Simon MA, Tseng CW, Wong JB. Risk assessment, genetic counseling, and genetic testing for BRCA-related Cancer: US Preventive Services Task Force Recommendation Statement. JAMA. 2019; 322(7):652-65.
Slade D. PARP and PARG inhibitors in cancer treatment. Genes Dev. 2020;34(5–6):360–94.
Dziadkowiec KN, Gąsiorowska E, Nowak-Markwitz E, Jankowska A. PARP inhibitors: review of mechanisms of action and BRCA1/2 mutation targeting. Prz Menopauzalny. 2016;15(4):215–9.
Rose M, Burgess JT, O’Byrne K, Richard DJ, Bolderson E. PARP inhibitors: clinical relevance, mechanisms of action and tumor resistance. Front Cell Dev Biol. 2020;8:564601.
Yi M, Dong B, Qin S, Chu Q, Wu K, Luo S. Advances and perspectives of PARP inhibitors. Exp Hematol Oncol. 2019;8:29.
Won KA, Spruck C. Triple-negative breast cancer therapy: current and future perspectives (Review). Int J Oncol. 2020;57(6):1245–61.
Huang CC, Tsai YF, Liu CY, Lien PJ, Wang YL, Lin YS, Chao TC, Feng CJ, Chiu JH, Chau GY, Tseng LM. Mutational profiles of BRCA 1/2 among Taiwanese Breast Cancers: Preliminary Results of 100 Consecutive Targeted Sequencing Samples. 2020 annual meeting of Taiwan Surgical Association.
Zhong X, Dong Z, Dong H, Li J, Peng Z, Deng L, Zhu X, Sun Y, Lu X, Shen F, Su X, Zhang L, Gu Y, Zheng H. Prevalence and prognostic role of BRCA1/2 variants in unselected Chinese breast cancer patients. PLoS One. 2016;11(6):e0156789.
Armstrong N, Ryder S, Forbes C, Ross J, Quek RG. A systematic review of the international prevalence of BRCA mutation in breast cancer. Clin Epidemiol. 2019;11(11):543–61.
Turner N, Tutt A, Ashworth A. Hallmarks of “BRCAness” in sporadic cancers. Nat Rev Cancer. 2004;4:814–9.
Wengner AM, Scholz A, Haendler B. Targeting DNA damage response in prostate and breast cancer. Int J Mol Sci. 2020;21(21):8273.
Huang CC, Tsai YF, Liu CY, Chao TC, Lien PJ, Lin YS, Feng CJ, Chiu JH, Hsu CY, Tseng LM. Comprehensive molecular profiling of Taiwanese breast cancers revealed potential therapeutic targets: prevalence of actionable mutations among 380 targeted sequencing analyses. BMC Cancer. 2021;21(1):199.
Burstein HJ, Curigliano G, Loibl S, Dubsky P, Gnant M, Poortmans P, Colleoni M, Denkert C, Piccart-Gebhart M, Regan M, Senn HJ, Winer EP, Thurlimann B. Members of the St. Gallen International consensus panel on the primary therapy of early breast cancer 2019. Estimating the benefits of therapy for early-stage breast cancer: the St. Gallen International Consensus Guidelines for the primary therapy of early breast cancer 2019. Ann Oncol. 2019;30(10):1541–57.
Curigliano G, Burstein HJ, Winer EP, Gnant M, Dubsky P, Loibl S, Colleoni M, Regan MM, Piccart-Gebhart M, Senn HJ, Thürlimann B; St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2017, André F, Baselga J, Bergh J, Bonnefoi H, Brucker SY, Cardoso F, Carey L, Ciruelos E, Cuzick J, Denkert C, Di Leo A, Ejlertsen B, Francis P, Galimberti V, Garber J, Gulluoglu B, Goodwin P, Harbeck N, Hayes DF, Huang CS, Huober J, Hussein K, Jassem J, Jiang Z, Karlsson P, Morrow M, Orecchia R, Osborne KC, Pagani O, Partridge AH, Pritchard K, Ro J, Rutgers EJT, Sedlmayer F, Semiglazov V, Shao Z, Smith I, Toi M, Tutt A, Viale G, Watanabe T, Whelan TJ, Xu B. De-escalating and escalating treatments for early-stage breast cancer: the St. Gallen International Expert Consensus Conference on the Primary Therapy of Early Breast Cancer 2017. Ann Oncol. 2017;28(8):1700-12.
Telli ML, Gradishar WJ, Ward JH. NCCN Guidelines updates: breast cancer. J Natl Compr Canc Netw. 2019;17(5.5):552-5.
Heeke AL, Pishvaian MJ, Lynce F, Xiu J, Brody JR, Chen WJ, Baker TM, Marshall JL, Isaacs C. Prevalence of homologous recombination-related gene mutations across multiple cancer types. JCO Precis Oncol. 2018;2018:PO.17.00286.
Gruber JJ, Afghahi A, Hatton A, Scott D, McMillan A, Ford JM, Telli ML. Talazoparib beyond BRCA: a phase II trial of talazoparib monotherapy in BRCA1 and BRCA2 wild-type patients with advanced HER2-negative breast cancer or other solid tumors with a mutation in homologous recombination (HR) pathway genes. J Clin Oncol. 2019;37(15_suppl):3006-6.
Landrum MJ, Chitipiralla S, Brown GR, Chen C, Gu B, Hart J, Hoffman D, Jang W, Kaur K, Liu C, Lyoshin V, Maddipatla Z, Maiti R, Mitchell J, O’Leary N, Riley GR, Shi W, Zhou G, Schneider V, Maglott D, Holmes JB, Kattman BL. ClinVar: improvements to accessing data. Nucleic Acids Res. 2020;48(D1):D835–44.
Lee CY, Chattopadhyay A, Chiang LM, Juang JJ, Lai LC, Tsai MH, Lu TP, Chuang EY. VariED: the first integrated database of gene annotation and expression profiles for variants related to human diseases. Database (Oxford). 2019;2019:baz075.
Jennings LJ, Arcila ME, Corless C, Kamel-Reid S, Lubin IM, Pfeifer J, Temple-Smolkin RL, Voelkerding KV, Nikiforova MN. Guidelines for validation of next-generation sequencing-based oncology panels: a joint consensus recommendation of the association for molecular pathology and college of American Pathologists. J Mol Diagn. 2017;19(3):341–65.
Dehghani M, Rosenblatt KP, Li L, Rakhade M, Amato RJ. Validation and clinical applications of a comprehensive next generation sequencing system for molecular characterization of solid cancer tissues. Front Mol Biosci. 2019;25(6):82.
Li MM, Datto M, Duncavage EJ, Kulkarni S, Lindeman NI, Roy S, Tsimberidou AM, Vnencak-Jones CL, Wolff DJ, Younes A, Nikiforova MN. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a Joint Consensus Recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn. 2017;19(1):4–23.
Oscanoa J, Sivapalan L, Gadaleta E, Dayem Ullah AZ, Lemoine NR, Chelala C. SNPnexus: a web server for functional annotation of human genome sequence variation (2020 update). Nucleic Acids Res. 2020;48(W1):W185–92.
Gonçalves A, Bertucci A, Bertucci F. Cancers (Basel). PARP inhibitors in the treatment of early breast cancer: the step beyond? 2020;12(6):1378.
Jalloul N, Gomy I, Stokes S, Gusev A, Johnson BE, Lindeman NI, Macconaill L, Ganesan S, Garber JE, Khiabanian H. Germline testing data validate inferences of mutational status for variants detected from tumor-only sequencing. JCO Precis Oncol. 2021;5:PO.21.00279.
Mateo J, Chakravarty D, Dienstmann R, Jezdic S, Gonzalez-Perez A, Lopez-Bigas N, Ng CKY, Bedard PL, Tortora G, Douillard JY, Van Allen EM, Schultz N, Swanton C, André F, Pusztai L. A framework to rank genomic alterations as targets for cancer precision medicine: the ESMO scale for clinical actionability of molecular targets (ESCAT). Ann Oncol. 2018;29(9):1895–902.
De Talhouet S, Peron J, Vuilleumier A, Friedlaender A, Viassolo V, Ayme A, Bodmer A, Treilleux I, Lang N, Tille JC, Chappuis PO, Buisson A, Giraud S, Lasset C, Bonadona V, Trédan O, Labidi-Galy SI. Clinical outcome of breast cancer in carriers of BRCA1 and BRCA2 mutations according to molecular subtypes. Sci Rep. 2020;10(1):7073.
Rennert G, Bisland-Naggan S, Barnett-Griness O, Bar-Joseph N, Zhang S, Rennert HS, Narod SA. Clinical outcomes of breast cancer in carriers of BRCA1 and BRCA2 mutations. N Engl J Med. 2007;357(2):115–23.
Lord CJ, Ashworth A. PARP inhibitors: synthetic lethality in the clinic. Science. 2017;355(6330):1152–8.
Robson M, Im SA, Senkus E, Xu B, Domchek SM, Masuda N, Delaloge S, Li W, Tung N, Armstrong A, Wu W, Goessl C, Runswick S, Conte P. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N Engl J Med. 2017;377(6):523–33.
Hurvitz SA, Gonçalves A, Rugo HS, Lee KH, Fehrenbacher L, Mina LA, Diab S, Blum JL, Chakrabarti J, Elmeliegy M, DeAnnuntis L, Gauthier E, Czibere A, Tudor IC, Quek RGW, Litton JK, Ettl J. Talazoparib in patients with a germline BRCA-mutated advanced breast cancer: detailed safety analyses from the phase III EMBRACA trial. Oncologist 2020;25:e439–50.
Heeke AL, Xiu J, Elliott A, Korn WM, Lynce F, Pohlmann PR, Isaacs C, Swain SM, Vidal G, Schwartzberg LS, Tan AR. Actionable coalterations in breast tumors with pathogenic mutations in the homologous recombination DNA damage repair pathway. Breast Cancer Res Treat. 2020;184(2):265–75.
Pellegrino B, Mateo J, Serra V, Balmaña J. Controversies in oncology: are genomic tests quantifying homologous recombination repair deficiency (HRD) useful for treatment decision making? ESMO Open. 2019;4(2):e000480.
Tung NM, Robson ME, Ventz S, Santa-Maria CA, Nanda R, Marcom PK, Shah PD, Ballinger TJ, Yang ES, Vinayak S, Melisko M, Brufsky A, DeMeo M, Jenkins C, Domchek S, D’Andrea A, Lin NU, Hughes ME, Carey LA, Wagle N, Wulf GM, Krop IE, Wolff AC, Winer EP, Garber JE. TBCRC 048: phase II study of Olaparib for metastatic breast cancer and mutations in homologous recombination-related genes. J Clin Oncol. 2020;38(36):4274–82.
VIOLETTE: a randomized phase II study to assess the DNA damage response inhibitors AZD6738 or AZD1775 in combination with olaparib (Ola) versus Ola monotherapy in patients (pts) with metastatic, triple-negative breast cancer (TNBC). J Clin Oncol. 2019;37(15_suppl):TPS1112.
Dougherty BA, Lai Z, Hodgson DR, Orr MCM, Hawryluk M, Sun J, Yelensky R, Spencer SK, Robertson JD, Ho TW, Fielding A, Ledermann JA, Barrett JC. Biological and clinical evidence for somatic mutations in BRCA1 and BRCA2 as predictive markers for olaparib response in high-grade serous ovarian cancers in the maintenance setting. Oncotarget. 2017;8(27):43653–61. https://doi.org/10.18632/oncotarget.17613.
Konstantinopoulos PA, Norquist B, Lacchetti C, Armstrong D, Grisham RN, Goodfellow PJ, Kohn EC, Levine DA, Liu JF, Lu KH, Sparacio D, Annunziata CM. Germline and somatic tumor testing in epithelial ovarian cancer: ASCO guideline. J Clin Oncol. 2020;38(11):1222–45.
Vikas P, Borcherding N, Chennamadhavuni A, Garje R. Therapeutic potential of combining PARP inhibitor and immunotherapy in solid tumors. Front Oncol. 2020;10:570.
Sharma P, Rodler E, Barlow WE, Gralow J, Huggins-Puhalla SL, Anders CK, Goldstein LJ, Brown-Glaberman UA, Huynh T, Szyarto CS, Godwin AK, Pathak HB, Swisher EM, Radke MR, Timms KM, Lew DL, Miao J, Pusztai L, Hayes DF, Hortobagyi GN. The results of a phase II randomized trial of cisplatin ± veliparib in metastatic triple-negative breast cancer (TNBC) and/or germline BRCA-associated breast cancer (SWOG S1416). J Clin Oncol. 2020;38(15_suppl):1001.
Telli ML, Timms KM, Reid J, Hennessy B, Mills GB, Jensen KC, Szallasi Z, Barry WT, Winer EP, Tung NM, Isakoff SJ, Ryan PD, Greene-Colozzi A, Gutin A, Sangale Z, Iliev D, Neff C, Abkevich V, Jones JT, Lanchbury JS, Hartman AR, Garber JE, Ford JM, Silver DP, Richardson AL. Homologous recombination deficiency (HRD) score predicts response to platinum-containing neoadjuvant chemotherapy in patients with triple-negative breast cancer. Clin Cancer Res. 2016;22(15):3764–73.
Clark DF, Maxwell KN, Powers J, Lieberman DB, Ebrahimzadeh J, Long JM, McKenna D, Shah P, Bradbury A, Morrissette JJD, Nathanson KL, Domchek SM. Identification and confirmation of potentially actionable germline mutations in tumor-only genomic sequencing. JCO Precis Oncol. 2019. https://doi.org/10.1200/PO.19.00076
Watkins JA, Irshad S, Grigoriadis A, Tutt AN. Genomic scars as biomarkers of homologous recombination deficiency and drug response in breast and ovarian cancers. Breast Cancer Res. 2014;16(3):211.
Gou R, Dong H, Lin B. Application and reflection of genomic scar assays in evaluating the efficacy of platinum salts and PARP inhibitors in cancer therapy. Life Sci. 2020;261:118434.
Tutt ANJ, Garber JE, Kaufman B, Viale G, Fumagalli D, Rastogi P, Gelber RD, De Azambuja E, Fielding A, Balmaña J, Domchek SM, Gelmon KA, Hollingsworth SJ, Korde LA, Linderholm B, Bandos H, Senkus E, Suga JM, Shao Z, Pippas AW, Nowecki Z, Huzarski T, Ganz PA, Lucas PC, Baker N, Loibl S, McConnell R, Piccart M, Schmutzler R, Steger GG, Costantino JP, Arahmani A, Wolmark N, McFadden E, Karantza V, Lakhani SR, Yothers G, Campbell C, Geyer CE. (2021) OlympiA Clinical Trial Steering Committee and Investigators. Adjuvant Olaparib for patients with BRCA1- or BRCA-mutated breast cancer. N Engl J Med. 2021; 384 (25): 2394-405.
Acknowledgment
The study was exclusively sponsored by YongLin Healthcare Foundation under clinical study protocol No. QCR18002. The funder had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. This manuscript contained materials presented during the Taipei International Breast Cancer Symposium 2020 Meeting during November 7 and 8, 2020. The authors would like to thank Dr. Morris Chang (ABMRD002) and the Melissa Lee Cancer Foundation for their help during the study period.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Disclosure
All authors declare that there are no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Huang, CC., Tsai, YF., Liu, CY. et al. Prevalence of Tumor Genomic Alterations in Homologous Recombination Repair Genes Among Taiwanese Breast Cancers. Ann Surg Oncol 29, 3578–3590 (2022). https://doi.org/10.1245/s10434-022-11347-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1245/s10434-022-11347-0