Abstract
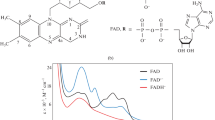
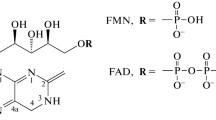
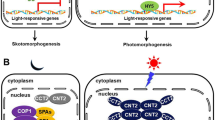
The blue-light sensors, cryptochromes, compose the extensive class of flavoprotein photoreceptors, regulating signaling processes in plants underlying their development, growth, and metabolism. In several algae, cryptochromes may act not only as sensory photoreceptors but also as photolyases, catalyzing repair of the UV-induced DNA lesions. Cryptochromes bind FAD as the chromophore at the photolyase homologous region (PHR) domain and contain the cryptochrome C-terminal extension (CCE), which is absent in photolyases. Photosensory process in cryptochrome is initiated by photochemical chromophore conversions, including formation of the FAD redox forms. In the state with the chromophore reduced to neutral radical (FADH•), the photoreceptor protein undergoes phosphorylation, conformational changes, and disengagement from the PHR domain and CCE with subsequent formation of oligomers of cryptochrome molecules. Photooligomerization is a structural basis of the functional activities of cryptochromes, since it ensures formation of their complexes with a variety of signaling proteins, including transcriptional factors and regulators of transcription. Interactions in such complexes change the protein signaling activities, leading to regulation of gene expression and plant photomorphogenesis. In recent years, multiple papers, reporting novel, more detailed information about the molecular mechanisms of above-mentioned processes were published. The present review mainly focuses on analysis of the data contained in these publications, particularly regarding structural aspects of the cryptochrome transitions into photoactivated states and regulatory signaling processes mediated by the cryptochrome photoreceptors in plants.




Similar content being viewed by others
Abbreviations
- 6-4PP:
-
pyrimidine 6-4 pyrimidone photoproduct
- BICs:
-
blue light inhibitors of CRYs
- CCE:
-
cryptochrome C-terminal extension
- CIB:
-
CRY-interacting bHLH
- CO:
-
constans
- COP1:
-
constitutive photomorphogenic 1
- CPD:
-
cyclobutane pyrimidine dimer
- CPH1:
-
Chlamydomonas photolyase homologous 1 plant-like pCRY of Chlamydomonas reinhardtii
- CPF:
-
cryptochrome/photolyase family
- CraCRY:
-
animal-like aCRY of C. reinhardtii
- CRY:
-
cryptochrome
- PIF:
-
phytochrome-interacting factor
- PPKs:
-
photoregulatory protein kinases
- LRGs:
-
light-responsive genes
- PHR:
-
photolyase homologous region
- SPA:
-
suppressor of PHYA-105 1
- UVB/UVA:
-
UV of B-region (290-320 nm)/UV of A-region (320-400 nm)
- UVR8:
-
UV resistance locus 8
References
Losi, A., and Gartner, W. (2012) The evolution of flavin-binding photoreceptors: an ancient chromophore serving trendy blue-light sensors, Annu. Rev. Plant Biol., 63, 49-72, https://doi.org/10.1146/annurev-arplant-042811-105538.
Fraikin, G. Ya., Strakhovskaya, M. G., and Rubin, A. B. (2013) Biological photoreceptors of light-dependent regulatory processes, Biochemistry (Moscow), 78, 1238-1253, https://doi.org/10.1134/S0006297913110047.
Li, F.-W., and Mathews, S. (2016) Evolutionary aspects of plant photoreceptors, J. Plant Res., 129, 115-122, https://doi.org/10.1007/s10265-016-0785-4.
Podolec, R., Demarsy, E., and Ulm, R. (2021) Perception and signaling of ultraviolet-B in plants, Annu. Rev. Plant Biol., 72, 793-822, https://doi.org/10.1146/annurev-arplant-050718-095946.
Inoue, K., Nishihama, R., and Kohchi, T. (2017) Evolutionary origin of phytochrome responses and signaling in land plants: phytochrome response and signaling in basal land plants, Plant Cell Environ., 40, 2502-2508, https://doi.org/10.1111/pce.12908.
Fraikin, G. Ya., Strakhovskaya, M. G., Belenikina, N. S., and Rubin, A. B. (2015) Bacterial photosensory proteins: regulatory functions and optogenetic applications, Microbiology, 84, 461-472, https://doi.org/10.1134/S0026261715040086.
Demarsy, E., Goldschmidt-Clemont, M., and Ulm, R. (2018) Coping with “dark sides of the sun” through photoreceptor signaling, Trends Plant Sci., 23, 260-271, https://doi.org/10.1016/j.tplants.2017.11.007.
Fraikin, G. Ya. (2018) Protein light sensors: photoexcited states, signaling properties and applications in optogenetics [in Russian], AR-Consalt, Moscow.
Ponnu, J., and Hoecker, U. (2022) Signaling mechanisms by Arabidopsis cryptochromes, Front. Plant Sci., 13, 844714, https://doi.org/10.3389/fpls.2022.844714.
Vechtomova, Y. L., Telegina, T. A., and Kritsky, M. S. (2020) Evolution of proteins of the DNA photolyase/cryptochrome family, Biochemistry (Moscow), 85 (Suppl. 1), S131-S153, https://doi.org/10.1134/S0006297920140072.
Wang, Q., and Lin, C. (2020) Mechanisms of cryptochrome-mediated photoresponses in plants, Annu. Rev. Plant Biol., 71, 103-129, https://doi.org/10.1146/annurev-arplant-050718-100300.
Fraikin, G. (2017) Photobioregulatory Receptors, Lambert Academic Publishing, Saarbrucken.
Sun, K., and Zhu, Z. (2018) Illuminating the nucleus: UVR8 interacts with more, Trends Plant Sci., 23, 279-281, https://doi.org/10.1016/j.tplants.2018.03.002.
Ahmad, M. (2016) Photocycle and signaling mechanisms of plant cryptochromes, Curr. Opin. Plant Biol., 33, 108-115, https://doi.org/10.1016/j.pbi.2016.06.013.
Christie, J. M., Blackwood, L., Petersen, J., and Sullivan, S. (2015) Plant flavoprotein photoreceptors, Plant Cell Physiol., 56, 401-413, https://doi.org/10.1093/pcp/pcu196.
Fraikin, G. Ya., and Rubin, A. B. (2022) Institute for Computer Research, in Horizons of Biophysics [in Russian] (Rubin, A. B., ed) Vol. 1, Moscow-Izhevsk, pp. 426-454.
Cheng, M. C., Kathare, P. K., Paik, I., and Hug, E. (2021) Phytochrome signaling networks, Annu. Rev. Plant Biol., 72, 217-244, https://doi.org/10.1146/annurev-arplant-080620-024221.
Bae, G., and Choi, G. (2008) Decoding of light signals by plant phytochromes and their interacting proteins, Annu. Rev. Plant Biol., 59, 281-311, https://doi.org/10.1146/annurev.arplant.59.032607.092859.
Franklin, K. A., and Quail, P. H. (2010) Phytochrome functions in Arabidopsis development, J. Exp. Bot., 61, 11-24, https://doi.org/10.1093/jxb/erp304.
Fraser, D. P., Hayes, S., and Franklin, K. A. (2016) Photoreceptor crosstalk in shade avoidance, Curr. Opin. Plant Biol., 33, 1-7, https://doi.org/10.1016/jpbi.2016.03.008.
Chen, M., and Chory, J. (2011) Phytochrome signaling mechanisms and the control of plant development, Trends Cell Biol., 21, 664-671, https://doi.org/10.1016/j.tcb.2011.07.002.
Pham, V. N., Kathare, P. K., and Hug, E. (2018) Phytochromes and phytochrome interacting factors, Plant Physiol., 176, 1025-1038, https://doi.org/10.1104/pp.17.01384.
Wang, Q., Zuo, Z., Wang, X., Liu, Q., Gu, L., Oka, Y., and Lin, C. (2018) Beyond the photocycle – how cryptochromes regulate photoresponses in plants? Curr. Opin. Plant Biol., 45, 120-126, https://doi.org/10.1016/j.pbi.2018.05.014.
Demarsy, E., and Fankhauser, C. (2009) Higher plants use LOV to perceive blue light, Curr. Opin. Plant Biol., 12, 69-74, https://doi.org/10.1016/j.pbi.2008.09.002.
Christie, J. M., Arvai, A. S., Baxter, K. J., Heilmann, M., Pratt, A. J., O’Hara, A., Kelly, S. M., Hothorn, M., Smith, B. O., Hitomi, K., Jenkins, G. I., and Getzoff, E. D. (2012) Plant UVR8 photoreceptor senses UV-B by tryptophan-mediated disruption of cross-dimer salt bridges, Science, 335, 1492-1496, https://doi.org/10.1126/science.1218091.
Fraikin, G. Ya. (2018) Signaling mechanisms regulating diverse plant cell responses to UVB radiation, Biochemistry (Moscow), 83, 787-794, https://doi.org/10.1134/S0006297918070027.
Rockwell, N. C., Shang, L., Martin, S. S., and Lagarias, J. C. (2009) Distinct classes of red/far-red photochemistry within the phytochrome superfamily, Proc. Natl. Acad. Sci. USA, 106, 6123-6127, https://doi.org/10.1073/pnas.0902370106.
Schwinn, K., Ferre, N., and Huix-Rotllant, M. (2020) UV-visible absorption spectrum of FAD and its reduced forms embedded in a cryptochrome protein, Phys. Chem. Chem. Phys., 22, 12447-12455, https://doi.org/10.1039/d0cp01714k.
Fraikin, G. Ya. (2022) Photosensory and signaling properties of cryptochromes, Mosc. Univ. Biol. Sci. Bull., 77, 54-63, https://doi.org/10.3103/S0096392522020031.
Ahmad, M., and Cashmore, A. R. (1993) HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor, Nature, 366, 162-166, https://doi.org/10.1038/366162a0.
Sancar, A. (2004) Photolyase and cryptochrome blue-light photoreceptors, Adv. Protein Chem., 69, 73-100, https://doi.org/10.1016/S0065-3233(04)69003-6.
Cashmore, A. R., Jarillo, J. A., Wu, Y. J., and Liu, D. (1999) Cryptochromes: blue light receptors for plants and animals, Science, 284, 760-765, https://doi.org/10.1126/science.284.5415.760.
Muller, M., and Carell, T. (2009) Structural biology of DNA photolyases and cryptochromes, Curr. Opin. Struct. Biol., 19, 277-285, https://doi.org/10.1016/j.sbi.2009.05.003.
Zoltowski, B. D. (2015) Resolving cryptic aspects of cryptochrome signaling, Proc. Natl. Acad. Sci. USA, 112, 8811-8812, https://doi.org/10.1073/pnas.1511092112.
Takahashi, J. S. (2017) Transcriptional architecture of the mammalian circadian clock, Nat. Rev. Genet., 18, 164-179, https://doi.org/10.1038/nrg.2016.150.
Wu, G., and Spalding, E. P. (2007) Separate functions for nuclear and cytoplasmic cryptochrome 1 during photomorphogenesis of Arabidopsis seedlings, Proc. Natl. Acad. Sci. USA, 104, 18813-18818, https://doi.org/10.1073/pnas.0705082104.
Yu, X., Klejnot, J., Zhao, X., Shalitin, D., Maymon, M., Yang, H., Lee, J., Liu, X., Lopez, J., and Lin, C. (2007) Arabidopsis cryptochrome 2 completes its posttranslational life cycle in the nucleus, Plant Cell, 19, 3146-3156, https://doi.org/10.1105/tpc.107.053017.
Pooam, M., Arthaut, L.-D., Burdick, D., Link, J., Martino, C. F., and Ahmad, M. (2019) Magnetic sensitivity mediated by the Arabidopsis blue-light receptor cryptochrome occurs during flavin reoxidation in the dark, Planta, 249, 319-332, https://doi.org/10.1007/s00425-018-3002-y.
Zoltowski, B. D., Chelliah, Y., Wickramaratne, A., Jarecha, L., Karki, N., Xu, W., Mouritsen, H., Hore, P. J., Hibbs, R. E., Green, C. B., and Takahashi, J. S. (2019) Chemical and structural analysis of a photoactive vertebrate cryptochrome from pigeon, Proc. Natl. Acad. Sci. USA, 116, 19449-19457, https://doi.org/10.1073/pnas.1907875116.
Petersen, J., Rredhi, A., Szyttenholm, J., Oldemeyer, S., Kottke, T., and Mittag, M. (2021) The world of algae reveals a broad variety of cryptochrome properties and functions, Front. Plant Sci., 12, 748760, https://doi.org/10.3389/fpls.2021.766509.
Palayam, M., Ganapathy, J., Guercio, A. M., Tal, L., Deck, S. L., and Shabek, K. N. (2021) Structural insights into photoactivation of plant cryptochrome-2, Commun. Biol., 4, 28, https://doi.org/10.1038/s42003-020-01531-x.
Chaves, I., Pokorny, R., Byrdin, M., Hoang, N., Ritz, T., Brettel, K., Essen, L.-O., van der Horst, G. T., Batschauer, A., and Ahmad, M. (2011) The cryptochromes: blue light photoreceptors in plants and animals, Annu. Rev. Plant Biol., 62, 335-364, https://doi.org/10.1146/annurev-arplant-042110-103759.
Ozturk, N. (2017) Phylogenetic and functional classification of the photolyase/cryptochrome family, Photochem. Photobiol., 93, 1-22, https://doi.org/10.1111/php.12676.
Zhang, M., Wang, L., and Zhong, D. (2017) Photolyase: dynamics and electron-transfer mechanisms of DNA repair, Arch. Biochem. Biophys., 632, 158-174, https://doi.org/10.1016/j.abb.2017.08.007.
Bayram, O., Braus, G. H., Fischer, R., and Rodriguez-Romero, J. (2010) Spotlight on Aspergillus nidulands photosensory systems, Fungal Genet. Biol., 47, 900-908, https://doi.org/10.1016/j.fgb.2010.05.008.
Konig, S., Juhas, M., Jager, S., Kottke, T., and Buchel, C. (2017) The cryptochrome-photolyase protein family in diatoms, J. Plant Physiol., 217, 15-19, https://doi.org/10.1016/j.plph.2017.06.015.
Kottke, T., Oldemeyer, S., Wenzel, S., Zou, Y., and Mittag, M. (2017) Cryptochrome photoreceptors in green algae: unexpected versatility of mechanisms and functions, J. Plant Physiol., 217, 4-14, https://doi.org/10.1016/j.plph.2017.05.021.
Michael, A. K., Fribourgh, J. L., Van Gelder, R. N., and Partch, C. L. (2017) Animal cryptochromes: divergent roles in light perception, circadian timekeeping and beyond, Photochem. Photobiol., 93, 128-140, https://doi.org/10.1111/php.12677.
Oldemeyer, S., Franz, S., Wenzel, S., Essen, L.-O., Mittag, M., and Kottke, T. (2016) Essential role of an unusual long-lived tyrosil radical in the response to red light of the animal-like cryptochrome aCRY, J. Biol. Chem., 291, 14062-14071, https://doi.org/10.1074/jbc.M116.726976.
Paulus, B., Bajzath, C., Melin, F., Heidinger, L., Kromm, V., Herkersdorf, C., Benz, U., Mann, L., Stehle, P., Hellwig, P., Weber, S., and Schleicher, E. (2015) Spectroscopic characterization of radicals and radical pairs in fruit fly cryptochrome – protonated and nonprotonated flavin radical-states, FEBS J., 282, 3175-3189, https://doi.org/10.1111/febs.13299.
Zoltowski, B. D., Vaidya, A. T., Top, D., Widom, J., Young, M. W., and Crane, B. R. (2011) Structure of full-length Drosophila cryptochrome, Nature, 480, 396-399, https://doi.org/10.1038/nature10618.
Sancar, A. (2008) Structure and function of photolyase and in vivo enzymology: 50th anniversary, J. Biol. Chem., 283, 32153-32157, https://doi.org/10.1074/jbc.R800052200.
Franz, S., Ignatz, E., Wenzel, S., Zielosko, H., Putu, E., Maestre-Reyna, M., Tsai, M.-D., Yamomoto, J., Mittag, M., and Essen, L.-O. (2018) Structure of the bifunctional cryptochrome aCRY from Chlamydomonas reinhardtii, Nucleic Acids Res., 46, 8010-8022, https://doi.org/10.1093/nar/gky621.
Hense, A., Herman, E., Oldemeyer, S., and Kottke, T. (2015) Proton transfer to flavin stabilizes the signaling state of the blue light receptor plant cryptochrome, J. Biol. Chem., 290, 1743-1751, https://doi.org/10.1074/jbc.M114.606327.
Lacombat, F., Espagne, A., Dozova, N., Plaza, P., Muller, P., Brettel, K., Franz-Badur, S., and Essen, L.-O. (2019) Ultrafast oxidation of a tyrosine by proton-coupled electron transfer promotes light activation of an animal-like cryptochrome, J. Am. Chem. Soc., 141, 13394-13409, https://doi.org/10.1021/jacs.9b03680.
Oldemeyer, S., Haddat, A. Z., and Fleming, G. R. (2020) Interconnection of the antenna pigment 8-HDF and flavin facilitates red-light reception in bifunctional animal-like cryptochrome, Biochemistry, 59, 594-604, https://doi.org/10.1021/acs.biochem.9b00875.
Goett-Zink, L., and Kottke, T. (2021) Plant cryptochromes illuminated: a spectroscopic perspective on the mechanism, Front. Chem., 9, 780199, https://doi.org/10.3389/fchem.2021.780199.
Thoing, C., Oldemeyer, S., and Kottke, T. (2015) Microsecond deprotonation of aspartic acid and response of the α/β subdomain precede C-terminal signaling in the blue light sensor plant cryptochrome, J. Am. Chem. Soc., 137, 5990-5999, https://doi.org/10.1021/jacs.5b01404.
Herbel, V., Orth, C., Wenzel, R., Ahmad, M., Bittl, R., and Batschauer, A. (2013) Lifetimes of Arabidopsis cryptochrome signaling states in vivo, Plant J., 74, 583-592, https://doi.org/10.1111/tpj.12144.
Muller, P., and Ahmad, M. (2011) Light-activated cryptochrome reacts with molecular oxygen to form a flavin-superoxide radical pair consistent with magnetoreception, J. Biol. Chem., 286, 21033-21040, https://doi.org/10.1074/jbc.M111.228940.
Goett-Zink, L., Toschke, A. L., Petersen, J., Mittag, M., and Kottke, T. (2021) C-terminal extension of plant cryptochrome dissociates from the β-sheet of the flavin-binding domain, J. Phys. Chem. Lett., 12, 5558-5563, https://doi.org/10.1021/acs.jpclett.1c00844.
Liu, Q., Su, T., He, W., Ren, H., Liu, S., Chen, Y., Gao, L., Hu, X., Lu, H., Cao, S., Huang, Y., Wang, X., Wang, Q., and Lin, C. (2020) Photooligomerization determines photosensitivity and photoreactivity of plant cryptochromes, Mol. Plant., 13, 398-413, https://doi.org/10.1016/j.molp.2020.01.002.
Yang, Z., Liu, B., Su, J., Liao, J., Lin, C., and Oka, Y. (2017) Cryptochromes orchestrate transcription regulation of diverse blue light responses in plants, Photochem. Photobiol., 93, 112-127, https://doi.org/10.1111/php.12663.
Wang, Q., and Lin, C. (2020) A structural view of plant CRY2 photoactivation and inactivation, Nat. Struct. Mol. Biol., 27, 401-403, https://doi.org/10.1038/s41594-020-0432-6.
Gao, J., Wang, X., Zhang, M., Bian, M., Deng, W., Zuo, Z., Yang, Z., Zhong, D., and Lin, C. (2015) Trp triad-dependent rapid photoreduction is not required for the function of Arabidopsis CRY1, Proc. Natl. Acad. Sci. USA, 112, 9135-9140, https://doi.org/10.1073/pnas.1504404112.
Liu, H., Su, T., He, W., Wang, G., and Lin, C. (2020) The universally conserved residues are not universally required for stable protein expression or functions of cryptochromes, Mol. Biol. Evol., 37, 327-340, https://doi.org/10.1093/molbev/msz217.
Shao, K., Zhang, X., Li, X., Hao, Y., Huang, X., Ma, M., Zhang, M., Yu, F., Liu, H., and Zhang, P. (2020) The oligomeric structures of plant cryptochromes, Nat. Struct. Mol. Biol., 27, 480-488, https://doi.org/10.1038/s41594-020-0420-x.
Wang, Q., Zuo, Z., Wang, X., Gu, L., Koshizumi, T., Yang, Z., Yang, L., Liu, Q., Liu, W., Han, Y. J., Kim, J. I., Liu, B., Wohlschlegel, J. A., Matsui, M., Oka, Y., and Lin, C. (2016) Photoactivation and inactivation of Arabidopsis cryptochrome 2, Science, 354, 343-347, https://doi.org/10.1126/science.aaf9030.
Ma, L., Wang, X., Guan, Z., Wang, L., Wang, Y., Zheng, L., Gong, Z., Shen, C., Wang, J., Zhang, D., Liu, Z., and Yin, P. (2020) Structural insight into BIC-mediated inactivation of Arabidopsis cryptochrome 2, Nat. Struct. Mol. Biol., 27, 472-479, https://doi.org/10.1038/s41594-020-0410-z.
Liu, H., Yu, X., Li, K., Klejnot, J., Yang, H., Lisiero, D., and Lin, C. (2008) Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis, Science, 322, 1535-1539, https://doi.org/10.1126/science.1163927.
Liu, Y., Li, X., Li, K., Lin, H., and Lin, C. (2013) Multiple bHLH proteins form heterodimers to mediate CRY2-dependent regulation of flowering-time in Arabidopsis, PLoS Genet., 9, e1003861, https://doi.org/10.1371/journal.pgen.1003861.
Liu, Y., Li, X., Ma, D., Chen, Z., Wang, J.-W., and Liu, H. (2018) CIB and CO interact to mediate CRY2-dependent regulation of flowering, EMBO Rep., 19, e45762, https://doi.org/10.15252/embr.2018.45762.
Ma, D., Li, X., Guo, Y., Chu, J., Fang, S., Yan, C., Noel, J. P., and Liu, H. (2016) Cryptochrome 1 interacts with PIFs to regulate high temperature-mediated hypocotyl elongation in response to blue light, Proc. Natl. Acad. Sci. USA, 113, 224-229, https://doi.org/10.1073/pnas.1511437113.
Pedmale, U. V., Huang, S. C., Zander, M., Cole, B. J., Herzel, J., Nery, J. R., and Ecker, J. R. (2016) Cryptochromes interact directly with PIFs to control plant growth in limiting blue light, Cell, 164, 233-245, https://doi.org/10.1016/j.cell.2015.12.018.
Ni, W., Xu, S.-L., Gonzalez-Grandio, E., Chalkley, R. J., Huhmer, A. F. R., Burlingame, A. L., Wang, Z.-Y., and Qual, P. H. (2017) PPKs mediate direct signal transfer from phytochrome photoreceptors to transcription factor PIF3, Nat. Commun., 8, 15236, https://doi.org/10.1038/ncomms15236.
Castillon, A., Shen, H., and Hug, E. (2009) Blue light induces degradation of the negative regulator phytochrome interacting factor 1 to promote photomorphogenic development of Arabidopsis seedlings, Genetics, 182, 161-171, https://doi.org/10.1534/genetics.108.099887.
Shalitin, D., Yang, H., Mockler, T. C., Maymon, M., Guo, H., Whitelam, G. C., and Lin, C. (2002) Regulation of Arabidopsis cryptochrome 2 by blue-light-dependent phosphorylation, Nature, 417, 763-767, https://doi.org/10.1038/nature00815.
Shalitin, D., Yu, X., Maymon, M., Mockler, T., and Lin, C. (2003) Blue-light-dependent in vivo and in vitro phosphorylation of Arabidopsis cryptochrome 1, Plant Cell, 15, 2421-2429, https://doi.org/10.1105/tpc.013011.
Weidler, G., zur Oven-Krockhaus, S., Heunemann, M., Orth, C., Schleifenbaum, F., Harter, K., Hoecker, U., and Batschauer, A. (2012) Degradation of Arabidopsis CRY2 is regulated by SPA proteins and phytochrome A, Plant Cell, 24, 2610-2623, https://doi.org/10.1105/tpc.112.098210.
Liu, Q., Wang, Q., Liu, B., Wang, W., Wang, X., Park, J., Yang, Z., Du, X., Bian, M., and Lin, C. (2016) The blue-light-dependent polyubiquitination and degradation of Arabidopsis cryptochrome 2 requires multiple E3 ubiquitin ligases, Plant Cell Physiol., 57, 2175-2186, https://doi.org/10.1093/pcp/pcw134.
Liu, Q., Wang, Q., Deng, W., Wang, X., Piao, M., Cai, D., Li, Y., Barshop, W.D., Yu, X., Zhou, T., Liu, B., Oka, Y., Wohlschlegel, J., Zuo, Z., and Lin, C. (2017) Molecular basis for blue light-dependent phosphorylation of Arabidopsis cryptochrome 2, Nat. Commun., 8, 15234, https://doi.org/10.1038/ncomms15234.
Wang, H., Ma, L. G., Li, G. M., Zhao, H. Y., and Deng, X. W. (2001) Direct interaction of Arabidopsis cryptochromes with COP1 in light control development, Science, 294, 154-158, https://doi.org/10.1126/science.1063630.
Yang, H.-Q., Tang, R.-H., and Cashmore, A. R. (2001) The signaling mechanism of Arabidopsis CRY1 involves direct interaction with COP1, Plant Cell, 13, 2573-2587, https://doi.org/10.1105/tpc.010367.
Ponnu, J. (2020) Molecular mechanisms suppressing COP1/SPA E3 ubiquitin ligase activity in blue light, Physiol. Plant, 169, 418-429, https://doi.org/10.1111/ppl.13103.
Heijde, M., and Ulm, R. (2012) UV-B photoreceptor-mediated signaling in plants, Trends Plant Sci., 17, 230-237, https://doi.org/10.1016/j.tplants.2012.01.007.
Hoecker, U. (2017) The activities of the E3 ubiquitin ligase COP1/SPA, a key repressor in light signaling, Curr. Opin. Plant Biol., 37, 63-69, https://doi.org/10.1016/j.pbi.2017.03.015.
Podolec, R., and Ulm, R. (2018) Photoreceptor-mediated regulation of the COP1/SPA E3 ubiquitin ligase, Curr. Opin. Plant Biol., 45, 18-25, https://doi.org/10.1016/j.pbi.2018.04.018.
Paik, I., Chen, F., Ngoc Pham, V., Zhu, L., Kim, J. I., and Hug, E. (2019) A phyB-PIF1-SPA1 kinase regulatory complex promotes photomorphogenesis in Arabidopsis, Nat. Commun., 10, 4216, https://doi.org/10.1038/s41467-019-12110-y.
Wang, W., Paik, I., Kim, j., Hou, X., Sung, S., and Hug, E. (2021) Direct phosphorylation of HY5 by SPA kinases to regulate photomorphogenesis in Arabidopsis, New Phytol., 230, 2311-2326, https://doi.org/10.1111/nph.17332.
Ponnu, J., and Hoecker, U. (2021) Illuminating the COP1/SPA ubiquitin ligase: fresh insight into its structure and functions during plant photomorphogenesis, Front. Plant Sci., 12, 662793, https://doi.org/10.3389/fpls.2021.662793.
Balzerowicz, M., Kemer, K., Schenkel, C., and Hoecker, U. (2017) SPA proteins affect the sub-cellular localization of COP1 in the COP1/SPA ubiquitin ligase complex during photomorphogenesis, Plant Physiol., 174, 1314-1321, https://doi.org/10.1104/pp.17.00488.
Kerner, K., Nagano, S., Lubbe, A., and Hoecker, U. (2021) Functional comparison of the WD-repeat domains of SPA1 and COP1 in suppression of photomorphogenesis, Plant Cell Environ., 44, 3273-3282, https://doi.org/10.1111/pce.14128.
Pacin, M., Legris, M., and Casal, J. J. (2014) Rapid decline in nuclear constitutive photomorphogenesis 1 abundance anticipates the stabilization of its target ELONGATED hypocotyl 5 in the light, Plant Physiol., 164, 1134-1138, https://doi.org/10.1104/pp.113.234245.
Lian, H. L., He, S. B., Zhang, Y. C., Zhu, D. M., Zhang, J. Y., Jia, K. P., San, S. X., Li, L., and Yang, H. Q. (2011) Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism, Genes Dev., 25, 1023-1028, https://doi.org/10.1101/gad.2025111.
Liu, B., Zuo, Z., Liu, H., Liu, X., and Lin, C. (2011) Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light, Genes Dev., 25, 1029-1034, https://doi.org/10.1101/gad.2025011.
Zuo, Z., Liu, H., Liu, B., Liu, X., and Lin, C. (2011) Blue-light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis, Curr. Biol., 21, 841-847, https://doi.org/10.1016/j.cub.2011.03.048.
Holtkotte, X., Ponnu, J., Ahmad, M., and Hoecker, U. (2017) The blue light-induced interaction of cryptochrome 1 with COP1 requires SPA proteins during Arabidopsis light signaling, PLoS Genet., 13, e1007044, https://doi.org/10.1371/journal.pgen.1007044.
Lau, K., Podolec, R., Chappuis, R., Ulm, R., and Hothorn, M. (2019) Plant photoreceptors and their signaling components compete for COP1 binding via VP peptide motifs, EMBO J., 38, e102140, https://doi.org/10.15252/embj.2019102140.
Ponnu, J., Riedel, T., Penner, E., Schrader, A., and Hoecker, U. (2019) Cryptochrome 2 competes with COP1-substrates to repress COP1 ligase activity during Arabidopsis photomorphogenesis, Proc. Natl. Acad. Sci. USA, 116, 27133-27141, https://doi.org/10.1073/pnas.1909181116.
Funding
This work was financially supported by the State Budget Assignment of Lomonosov Moscow State University, project no. 121032500058-7.
Author information
Authors and Affiliations
Contributions
G.Ya.F. – conceptualization, analysis of publications, writing and editing the text; N.S.B. – manuscript and figure preparation; A.B.R. – discussion of the manuscript material with coauthors.
Corresponding author
Ethics declarations
The authors declare no conflict of interest in financial or any other sphere. This article does not contain any studies with human participants or animals performed by any of the authors.
Rights and permissions
About this article
Cite this article
Fraikin, G.Y., Belenikina, N.S. & Rubin, A.B. Molecular Bases of Signaling Processes Regulated by Cryptochrome Sensory Photoreceptors in Plants. Biochemistry Moscow 88, 770–782 (2023). https://doi.org/10.1134/S0006297923060056
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0006297923060056