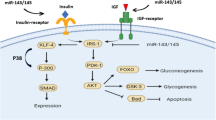
Abstract
Type 2 Diabetes Mellitus (T2DM) and cardiac hypertrophy (CH) are among the top ten leading cause of deaths, worldwide. T2DM and cardiac hypertrophy are the chronic diseases, have close association and direct life-threatening complications like stroke, myocardial infarction, retinopathy, nephropathy, and limb amputation. In addition to other medical approaches, miRNAs-based strategy is considered most efficient for early detection of chronic diseases and also has potential for the treatment of T2DM and cardiac hypertrophy like it is being used for cancer in clinical trials. MicroRNAs (miRNAs) are single stranded (non-coding) of 20 to 22 nucleotides sequences which bind to their target mRNA upon the complimentary basis, to silence the protein expression at post transcriptional level. Bioinformatic databases are used like online mendelian inheritance in man (OMIM), gene testing registry (GTR), TargetScan and ShinyGO for validation of disease linked genes and sorting the common miRNAs in both diseases, such as miR-30-5p/101-3p.2/190-5p/506-3p/9-5p/128-3p/137/96-5p/7-5p/107/101-3p.1/98-5p/124-3p.2/124-3p.116-5p/15-5p/497-5p/ 424-5p/195-5p/1271-5p, let-7-5p. Aforementioned databases were also used for the miRNAs which have more than one disease linked genes target in each pathological condition. Such miRNAs for cardiac hypertrophy are: miR-19-3p/183-5p.2/153-3p/372-3p/302-3p/520-3p/373-3p/129-5p/144-3p/139-5p and for T2DM are: miR-27-3p/206/1-3p/181-5p. This finding would be helpful for the appropriate selection of miRNAs and to design applicable research project in future. It will require more validation by using the miRNAs expression analysis, mimic, and anti-miRNA approach to check their potential against cardiac hypertrophy and T2DM.







Similar content being viewed by others
REFERENCES
WHO. 2019. https://www.who.int/news-room/fact-sheets/detail/the-top-10-causes-of-death. Accessed December 22, 2020.
Heron M. 2018. Leading causes for death. 2016. Natl. Vital. Stat. Rep. 67 (6), 1‒77.
Ahmad F.B., Anderson R.N.J.J. 2021. The leading causes of death in the US for 2020. JAMA. 325 (18), 1829‒1830.
American Diabetes Association. 2017. Standards of medical care in diabetes‒2017 abridged for primary care providers. Clin. Diabetes. 35 (1), 5‒26.
Olokoba A.B., Obateru O.A., Olokoba L.B. 2012. Type 2 diabetes mellitus: a review of current trends. Oman Med. J. 27 (4), 269‒273.
Nakamura M., Sadoshima J. 2018. Mechanisms of physiological and pathological cardiac hypertrophy. Nat. Rev. Cardiol. 15 (7), 387‒407.
Tham Y.K., Bernardo B.C., Ooi J.Y., Weeks K.L., McMullen J.R. 2015. Pathophysiology of cardiac hypertrophy and heart failure: signaling pathways and novel therapeutic targets. Arch. Toxicol. 89 (9), 1401‒1438.
Weeks K.L., McMullen J.R. 2011. The athlete’s heart vs. the failing heart: can signaling explain the two distinct outcomes. Physiol. J. 26 (2), 97‒105.
Laakso M. 2001. Cardiovascular disease in type 2 diabetes: challenge for treatment and prevention. J. Intern. Med. 249 (3), 225‒235.
Bartel D.P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 116 (2), 281‒297.
Ha T.Y. 2011. MicroRNAs in human diseases: from cancer to cardiovascular disease. Immune Netw.11 (3), 135‒154.
Wang J., Yang X. 2012.The function of miRNA in cardiac hypertrophy. Cell. Mol. Life Sci. 69 (21), 3561‒3570.
Smolarz B., Durczyński A., Romanowicz H., Szyłło K., Hogendorf P. 2022. miRNAs in cancer (review of literature). Int. J. Mol. Sci. 23 (5), 2805.
He B., Cai Q., Zhang Y., Zhang P., Shi S., Xie H., Peng X., Yin W., Tao Y., Wang X. 2020. miRNA-based biomarkers, therapies, and resistance in cancer. Int. J. Biol. Sci. 16 (14), 2628.
Pérez-Cremades D., Chen J. Assa C., Feinberg M.W. 2022. MicroRNA-mediated control of myocardial infarction in diabetes. Trends Cardiovasc. Med. S1050–1738(22)00006-8
Lavenniah A., Luu T.D., Li Y.P., Lim T.B., Jiang J., Ackers-Johnson M., Foo R.S. 2020. Engineered circular RNA sponges act as miRNA inhibitors to attenuate pressure overload-induced cardiac hypertrophy. Mol. Ther. 28 (6), 1506‒1517.
Zhou S.S., Jin J.P., Wang J.Q., Zhang Z.G., Freedman J.H., Zheng Y., Cai L. 2018. miRNAS in cardiovascular diseases: potential biomarkers, therapeutic targets and challenges. Acta Pharmacol. Sin. 39 (7), 1073‒1084.
Vasu S., Kumano K., Darden C.M., Rahman I., Lawrence M.C., Naziruddin B. 2019. MicroRNA signatures as future biomarkers for diagnosis of diabetes states. Cells. 8 (12), 1533.
Rubinstein W.S., Maglott D.R., Lee J.M., Kattman B.L., Malheiro A.J., Ovetsky M., Hem V., Gorelenkov V., Song G., Wallin C., Husain N. 2012. The NIH genetic testing registry: a new, centralized database of genetic tests to enable access to comprehensive information and improve transparency. Nucleic Acids Res. 41 (D1), D925‒D935.
Hamosh A., Scott A.F., Amberger J.S., Bocchini C.A., McKusick V.A. 2005. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 33 (D1), D514‒D517.
Agarwal V., Bell G.W., Nam J.W., Bartel D.P. 2015. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 4, e05005.
Dweep H., Sticht C., Pandey P., Gretz N. 2011. miRWalk–database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J. Biomed. Inform. 44 (5), 839‒847.
Griffiths-Jones S., Grocock R.J., Van Dongen S., Bateman A., Enright A.J. 2006. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 34 (D1), D140‒D144.
Ge S.X., Jung D., Yao R. 2020. ShinyGO: a graphical gene-set enrichment tool for animals and plants. J. Bioinform. 36 (8), 2628‒2629.
Zhang S. Cheng Z. Wang Y. Han T. 2021. The risks of miRNA therapeutics: in a drug target perspective. 15, 721.
Chiefari E., Nevolo M.T., Arcidiacono B., Maurizio E., Nocera A., Iiritano S., Sgarra R., Possidente K., Palmieri C., Paonessa F., Brunetti G. 2012. HMGA1 is a novel downstream nuclear target of the insulin receptor signaling pathway. Sci. Rep. 2 (1), 1‒10.
Hu Y., Li Q., Zhang L., Zhong L., Gu M., He B., Qu Q., Lao Y., Gu K., Zheng B., Yang H. 2021. Serum miR-195-5p exhibits clinical significance in the diagnosis of essential hypertension with type 2 diabetes mellitus by targeting DRD1. Clinics (Sao Paulo). 76, e2502. https://doi.org/10.6061/clinics/2021/e250228
Højlund K. 2014. Metabolism and insulin signaling in common metabolic disorders and inherited insulin resistance. Dan Med J. 61 (7), B4890.
Qian H.F., Li. Y., Wang L. 2017. Vaccinium bracteatum Thunb. Leaves’ polysaccharide alleviates hepatic gluconeogenesis via the downregulation of miR-137. Biomed. Pharmacother. 95, 1397-1403.
Wang D., Fang J., Lv J., Pan Z., Yin X., Cheng H., Guo X. 2019. Novel polymorphisms in PDLIM3 and PDLIM5 gene encoding Z-line proteins increase risk of idiopathic dilated cardiomyopathy. J. Cell. Mol. Med. 23 (10), 7054‒7062.
Elgebaly S.A., Christenson R.H., Kandil H., Ibrahim M., Rizk H., El-Khazragy N., Rashed L., Yacoub B., Eldeeb H., Ali M.M., Kreutzer D.L. 2021. Nourin-dependent miR-137 and miR-106b: novel biomarkers for early diagnosis of myocardial ischemia in coronary artery disease patients. Diagnostics (Basel). 11 (4), 703.
Einarson T.R., Acs A., Ludwig C., Panton U.H. 2018. Prevalence of cardiovascular disease in type 2 diabetes: a systematic literature review of scientific evidence from across the world in 2007–2017. Cardiovasc. Diabetol. 17 (1), 83.
Li J., Chattopadhyay K., Xu M., Chen Y., Hu F., Chu J., Li L. 2020. Prevalence and associated factors of vascular complications among inpatients with type 2 diabetes: a retrospective database study at a tertiary care department, Ningbo, China. PLoS One. 15 (6), e0235161.
Funding
We acknowledge the university research fund (URF) of Quaid-i-Azam University, Islamabad, Higher Education Commission (HEC) Pakistan, and Pakistan Science Foundation (PSF) to support the current study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no conflicts of interest.
This article does not contain any research involving animals or humans as subjects of research.
Rights and permissions
About this article
Cite this article
Hussain, K., Ishtiaq, A., Mushtaq, I. et al. Profiling of Targeted miRNAs (8-nt) for the Genes Involved in Type 2 Diabetes Mellitus and Cardiac Hypertrophy. Mol Biol 57, 338–345 (2023). https://doi.org/10.1134/S0026893323020085
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026893323020085