Abstract
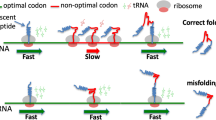
The genetic code sets the correspondence between the sequence of a given nucleotide triplet in an mRNA molecule, called a codon, and the amino acid that is added to the growing polypeptide chain during protein synthesis. With four bases (A, G, U, and C), there are 64 possible triplet codons: 61 sense codons (encoding amino acids) and 3 nonsense codons (so-called, stop codons that define termination of translation). In most organisms, there are 20 common/standard amino acids used in protein synthesis; thus, the genetic code is redundant with most amino acids (with the exception of Met and Trp) are being encoded by more than one (synonymous) codon. Synonymous codons were initially presumed to have entirely equivalent functions, however, the finding that synonymous codons are not present at equal frequencies in mRNA suggested that the specific codon choice might have functional implications beyond coding for amino acid. Observation of nonequivalent use of codons in mRNAs implied a possibility of the existence of auxiliary information in the genetic code. Indeed, it has been found that genetic code contains several layers of such additional information and that synonymous codons are strategically placed within mRNAs to ensure a particular translation kinetics facilitating and fine-tuning co-translational protein folding in the cell via step-wise/sequential structuring of distinct regions of the polypeptide chain emerging from the ribosome at different points in time. This review summarizes key findings in the field that have identified the role of synonymous codons and their usage in protein folding in the cell.




Similar content being viewed by others
Abbreviations
- ADAMTS13:
-
a disintegrin and metalloproteinase with a thrombospondin type 1 motif, member 13
- CAT:
-
chloramphenicol acetyltransferase
- FRET:
-
fluorescence resonance energy transfer
- mRNA:
-
messenger RNA
- NMR:
-
Nuclear magnetic resonance
- PGK:
-
phosphoglycerate kinase
- P-gp:
-
P-glycoprotein
- RNase A:
-
Ribonuclease A
- tRNA:
-
transport RNA
References
Crick, F. H., Barnett, L., Brenner, S., and Watts-Tobin, R. J. (1961) General nature of the genetic code for proteins, Nature, 192, 1227-1232.
Nirenberg, M., and Leder, P. (1964) RNA codewords and protein synthesis. The effect of trinucleotides upon the binding of sRNA to ribosomes, Science, 145, 1399-1407.
Leder, P., and Nirenberg, M. (1964) RNA codewords and protein synthesis. II. nucleotide sequence of a valine RNA codeword, Proc. Natl. Acad. Sci. USA, 52, 420-427.
Bernfield, M. R., and Nirenberg, M. W. (1965) RNA codewords and protein synthesis. The nucleotide sequences of multiple codewords for phenylalanine, serine, leucine, and proline, Science, 147, 479-484.
Trupin, J. S., Rottman, F. M., Brimacombe, R. L., Leder, P., Bernfield, M. R., and Nirenberg, M. W. (1965) RNA codewords and protein synthesis, VI. On the nucleotide sequences of degenerate codeword sets for isoleucine, tyrosine, asparagine, and lysine, Proc. Natl. Acad. Sci. USA, 53, 807-811.
Nirenberg, M., Leder, P., Bernfield, M., Brimacombe, R., Trupin, J., et al. (1965) RNA codewords and protein synthesis, VII. On the general nature of the RNA code, Proc. Natl. Acad. Sci. USA, 53, 1161-1168.
Brimacombe, R., Trupin, J., Nirenberg, M., Leder, P., Bernfield, M., and Jaouni, T. (1965) RNA codewords and protein synthesis, 8. Nucleotide sequences of synonym codons for arginine, valine, cysteine, and alanine, Proc. Natl. Acad. Sci. USA, 54, 954-960.
Söll, D., Ohtsuka, E., Jones, D. S., Lohrmann, R., Hayatsu, H., et al. (1965) Studies on polynucleotides, XLIX. Stimulation of the binding of aminoacyl-sRNA’s to ribosomes by ribotrinucleotides and a survey of codon assignments for 20 amino acids, Proc. Natl. Acad. Sci. USA, 54, 1378-1385.
Goel, N. S., Rao, G. S., Ycas, M., Bremermann, H. J., and King, L. (1972) A method for calculating codon frequencies in DNA, J. Theor. Biol., 35, 399-457.
Grantham, R., Gautier, C., Gouy, M., Mercier, R., and Pavé, A. (1980) Codon catalog usage and the genome hypothesis, Nucleic Acids Res., 8, r49-r62.
Grantham, R., Gautier, C., and Gouy, M. (1980) Codon frequencies in 119 individual genes confirm consistent choices of degenerate bases according to genome type, Nucleic Acids Res., 8, 1893-1912.
Ikemura, T. (1981) Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes, J. Mol. Biol., 146, 1-21.
Ikemura, T. (1982) Correlation between the abundance of yeast transfer RNAs and the occurrence of the respective codons in protein genes. Differences in synonymous codon choice patterns of yeast and Escherichia coli with reference to the abundance of isoaccepting transfer RNAs, J. Mol. Biol., 158, 573-597.
Bennetzen, J. L., and Hall, B. D. (1982) Codon selection in yeast, J. Biol. Chem., 257, 3026-3031.
Hastings, K. E. M., and Emerson, C. P. Jr. (1983) Codon usage in muscle genes and liver genes, J. Mol. Evol., 19, 214-218.
Ikemura, T. (1985) Codon usage and tRNA content in unicellular and multicellular organisms, Mol. Biol. Evol., 2, 13-34.
Sharp, P. M., Tuohy, T. M., and Mosurski, K. R. (1986) Codon usage in yeast: cluster analysis clearly differentiates highly and lowly expressed genes, Nucleic Acids Res., 14, 5125-5143.
Sharp, P. M., and Li, W. H. (1987) The Codon Adaptation Index – a measure of directional synonymous codon usage bias, and its potential applications, Nucleic Acids Res., 15, 1281-1295.
Sharp, P. M., Cowe, E., Higgins, D. G., Shields, D. C., Wolfe, K. H., and Wright, F. (1988) , Nucleic Acids Res., 16, 8207-8211.
Sørensen, M.A., Kurland, C. G., and Pedersen, S. (1989) Codon usage determines translation rate in Escherichia coli, J. Mol. Biol., 207, 365-377.
Andersson, S. G., and Kurland, C. G. (1990) Codon preferences in free-living microorganisms, Microbiol. Rev., 54, 198-210.
Kurland, C. G. (1991) Codon bias and gene expression, FEBS Lett., 285, 165-169.
Kramer, E. B., and Farabaugh, P. J. (2007) The frequency of translational misreading errors in E. coli is largely determined by tRNA competition, RNA, 13, 87-96.
Zaher, H. S., and Green, R. (2009) Fidelity at the molecular level: lessons from protein synthesis, Cell, 136, 746-762.
Kramer, E. B., Vallabhaneni, H., Mayer, L. M., and Farabaugh, P. J. (2010) A comprehensive analysis of translational missense errors in the yeast Saccharomyces cerevisiae, RNA, 16, 1797-1808.
Ribas de Pouplana, L., Santos, M. A., Zhu, J. H., Farabaugh, P. J., and Javid, B. (2014) Protein mistranslation: friend or foe?, Trends. Biochem. Sci., 39, 355-362.
Huang, Y., Koonin, E. V., Lipman, D. J., and Przytycka, T. M. (2009) Selection for minimization of translational frameshifting errors as a factor in the evolution of codon usage, Nucleic Acids Res., 37, 6799-6810.
Savir, Y., and Tlusty, T. (2013) The ribosome as an optimal decoder: a lesson in molecular recognition, Cell, 153, 471-479.
Dana, A., and Tuller, T. (2014) The effect of tRNA levels on decoding times of mRNA codons, Nucleic Acids Res., 42, 9171-9181.
Bonekamp, F., Andersen, H. D., Christensen, T., and Jensen, K. F. (1985) Codon-defined ribosomal pausing in Escherichia coli detected by using the pyrE attenuator to probe the coupling between transcription and translation, Nucleic Acids Res., 13, 4113-4123.
Curran, J. F., and Yarus, M. (1989) Rates of aminoacyl-tRNA selection at 29 sense codons in vivo, J. Mol. Biol., 209, 65-77.
Bulmer, M. (1987) Coevolution of codon usage and transfer RNA abundance, Nature, 325, 728-730.
Duret, L. (2000) tRNA gene number and codon usage in the C. elegans genome are co-adapted for optimal translation of highly expressed genes, Trends Genet., 16, 287-289.
Li, W. H. (1987) Models of nearly neutral mutation with particular implications for nonrandom usage of synonymous codons, J. Mol. Evol., 24, 337-345.
Shields, D. C. (1990) Switches in species-specific codon preferences: the influence of mutation biases, J. Mol. Evol., 31, 71-80.
Bulmer, M. (1991) The selection-mutation-drift theory of synonymous codon usage, Genetics, 129, 897-907.
Presnyak, V., Alhusaini, N., Chen, Y. H., Martin, S., Morris, N., et al. (2015) Codon optimality is a major determinant of mRNA stability, Cell, 160, 1111-1124, https://doi.org/10.1016/j.cell.2015.02.029.
Boël, G., Letso, R., Neely, H., Price, W. N., Wong, K. H., et al. (2016) Codon influence on protein expression in E. coli correlates with mRNA levels, Nature, 529, 358-363.
Mishima, Y., and Tomari, Y. (2016) Codon usage and 3′-UTR length determine maternal mRNA stability in zebrafish, Mol. Cell, 61, 874-885.
Bazzini, A. A., Del Viso, F., Moreno-Mateos, M. A., Johnstone, T. G., Vejnar, C. E., et al. (2016) Codon identity regulates mRNA stability and translation efficiency during the maternal-to-zygotic transition, EMBO J., 35, 2087-2103.
Radhakrishnan, A., Chen, Y. H., Martin, S., Alhusaini, N., Green, R., and Coller, J. (2016) The DEAD-box protein Dhh1p couples mRNA decay and translation by monitoring codon optimality, Cell, 167, 122-132.
Hia, F., Yang, S. F., Shichino, Y., Yoshinaga, M., Murakawa, Y., et al. (2019) Codon bias confers stability to human mRNAs, EMBO Rep., 20, e48220.
Wu, Q., Medina, S. G., Kushawah, G., DeVore, M. L., Castellano, L. A., et al. (2019) Translation affects mRNA stability in a codon-dependent manner in human cells, Elife, 8, e45396.
Medina-Muñoz, S. G., Kushawah, G., Castellano, L. A., Diez, M., DeVore, M. L., et al. (2021) Crosstalk between codon optimality and cis-regulatory elements dictates mRNA stability, Genome Biol., 22, 14.
Anfinsen, C. B. (1973) Principles that govern the folding of protein chains, Science, 181, 223-230.
Jaenicke, R. (1991) Protein folding: local structures, domains, subunits, and assemblies, Biochemistry, 30, 3147-3161.
Fersht, A. R. (2008) From the first protein structures to our current knowledge of protein folding: delights and skepticisms, Nat. Rev. Mol. Cell Biol., 9, 650-654.
Bartlett, A. I., and Radford, S. E. (2009) An expanding arsenal of experimental methods yields an explosion of insights into protein folding mechanisms, Nat. Struct. Mol. Biol., 16, 582-588.
Finkelstein, A. V. (2018) 50+ years of protein folding, Biochemistry (Moscow), 83 (Suppl. 1), S3-S18.
Abaskharon, R. M., and Gai, F. (2016) Meandering down the energy landscape of protein folding: are we there yet?, Biophys. J., 110, 1924-1932.
Ferina, J., and Daggett, V. (2019) Visualizing protein folding and unfolding, J. Mol. Biol., 431, 1540-1564.
Cowie, D. B., Spiegelman, S., Roberts, R. B., and Duerksen, J. D. (1961) Ribosome-bound β-galactosidase, Proc. Natl. Acad. Sci. USA, 47, 114-122.
Zipser, D., and Perrin, D. (1963) Complementation on ribosomes, Cold Spring Harb. Symp. Quant. Biol., 28, 533-537.
Kiho, Y., and Rich, A. (1964) Induced enzyme formed on bacterial polyribosomes, Proc. Natl. Acad. Sci. USA, 51, 111-118.
Hamlin, J., and Zabin, I. (1972) β-Galactosidase: immunological activity of ribosome-bound, growing polypeptide chains, Proc. Natl. Acad. Sci. USA, 69, 412-416.
Bergman, L. W., and Kuehl, W. M. (1979) Formation of intermolecular disulfide bonds on nascent immunoglobulin polypeptides, J. Biol. Chem., 254, 5690-5694.
Bergman, L. W., and Kuehl, W. M. (1979) Formation of an intrachain disulfide bond on nascent immunoglobulin light chains, J. Biol. Chem., 254, 8869-8876.
Bergman, L. W., and Kuehl, W. M. (1979) Co-translational modification of nascent immunoglobulin heavy and light chains, J. Supramol. Struct., 11, 9-24.
Lim, V. I., and Spirin, A. S. (1985) Stereochemistry of the transpeptidation reaction in the ribosome. The ribosome generates an alpha-helix in the synthesis of the protein polypeptide chain, Dokl. Akad. Nauk SSSR, 280, 235-239.
Lim, V. I., and Spirin, A. S. (1986) Stereochemical analysis of ribosomal transpeptidation. Conformation of nascent peptide, J. Mol. Biol., 188, 565-574.
Komar, A. A. (2019) Synonymous codon usage – a guide for co-translational protein folding in the cell, Mol. Biol. (Mosk), 53, 883-898.
Noé, F., De Fabritiis, G., and Clementi, C. (2020) Machine learning for protein folding and dynamics, Curr. Opin. Struct. Biol., 60, 77-84.
Gao, W., Mahajan, S. P., Sulam, J., and Gray, J. J. (2020) Deep learning in protein structural modeling and design, Patterns (NY), 1, 100142.
Senior, A. W., Evans, R., Jumper, J., Kirkpatrick, J., Sifre, L., et al. (2020) Improved protein structure prediction using potentials from deep learning, Nature, 577, 706-710.
Lindberg, M. O., and Oliveberg, M. (2007) Malleability of protein folding pathways: a simple reason for complex behaviour, Curr. Opin. Struct. Biol., 17, 21-29.
Englander, S. W., and Mayne, L. (2014) The nature of protein folding pathways, Proc. Natl. Acad. Sci. USA, 111, 15873-15880.
Englander, S. W., Mayne, L., Kan, Z. Y., and Hu, W. (2016) Protein folding-how and why: by hydrogen exchange, fragment separation, and mass spectrometry, Annu. Rev. Biophys., 45, 135-152.
Levinthal, C. (1969). How to fold graciously, in Mossbauer Spectroscopy in Biological Systems, Proceedings of a Meeting held at Allerton House, Monticello, Illinois (Debrunner, P., Tsibris, J. C. M., and Münck, E., eds.) University of Illinois Press, Urbana, p. 22.
Gopan, G., Gruebele, M., and Rickard, M. (2020) In-cell protein landscapes: making the match between theory, simulation and experiment, Curr. Opin. Struct. Biol., 66, 163-169.
Gershenson, A., Gosavi, S., Faccioli, P., and Wintrode, P. L. (2020) Successes and challenges in simulating the folding of large proteins, J. Biol. Chem., 295, 15-33.
Chow, M. K., Amin, A. A., Fulton, K. F., Fernando, T., Kamau, L., et al. (2006) The REFOLD database: a tool for the optimization of protein expression and refolding, Nucleic Acids Res., 34 (Database issue), D207-212.
Mizutani, H., Sugawara, H., Buckle, A. M., Sangawa, T., Miyazono, K. I., et al. (2017) REFOLDdb: a new and sustainable gateway to experimental protocols for protein refolding, BMC Struct. Biol., 17, 4.
Hartl, F. U., and Hayer-Hartl, M. (2009) Converging concepts of protein folding in vitro and in vivo, Nat. Struct. Mol. Biol., 16, 574-581.
Hingorani, K. S., and Gierasch, L. M. (2014) Comparing protein folding in vitro and in vivo: foldability meets the fitness challenge, Curr. Opin. Struct. Biol., 24, 81-90.
Balchin, D., Hayer-Hartl, M., and Hartl, F. U. (2016) In vivo aspects of protein folding and quality control, Science, 353, aac4354.
Gruebele, M., Dave, K., and Sukenik, S. (2016) Globular protein folding in vitro and in vivo, Annu. Rev. Biophys., 45, 233-251.
Dahiya, V., and Buchner, J. (2019) Functional principles and regulation of molecular chaperones, Adv. Protein. Chem. Struct. Biol., 114, 1-60.
Jayaraj, G. G., Hipp, M. S., and Hartl, F. U. (2019) Functional modules of the proteostasis network, Cold Spring Harb. Perspect. Biol., 12, a033951.
Komar, A. A. (2009) A pause for thought along the co-translational folding pathway, Trends Biochem. Sci., 34, 16-24.
Gloge, F., Becker, A. H., Kramer, G., and Bukau, B. (2014) Co-translational mechanisms of protein maturation, Curr. Opin. Struct. Biol., 24, 24-33.
Thommen, M., Holtkamp, W., and Rodnina, M. V. (2017) Co-translational protein folding: progress and methods, Curr. Opin. Struct. Biol., 42, 83-89.
Komar, A. A. (2018) Unraveling co-translational protein folding: concepts and methods, Methods, 137, 71-81.
Williams, N. K., and Dichtl, B. (2018) Co-translational control of protein complex formation: a fundamental pathway of cellular organization?, Biochem. Soc. Trans., 46, 197-206.
Waudby, C. A., Dobson, C. M., and Christodoulou, J. (2019) Nature and regulation of protein folding on the ribosome, Trends Biochem. Sci., 44, 914-926.
Jha, S., and Komar, A. A. (2011) Birth, life and death of nascent polypeptide chains, Biotechnol. J., 6, 623-640.
Das, D., Das, A., Samanta, D., Ghosh, J., Dasgupta, S., et al. (2008) Role of the ribosome in protein folding, Biotechnol. J., 3, 999-1009.
Voisset, C., Saupe, S. J., and Blondel, M. (2011) The various facets of the protein-folding activity of the ribosome, Biotechnol. J., 6, 668-673.
Banerjee, D., and Sanyal, S. (2014) Protein folding activity of the ribosome (PFAR) – a target for antiprion compounds, Viruses, 6, 3907-3924.
Woolhead, C. A., McCormick, P. J., and Johnson, A. E. (2004) Nascent membrane and secretory proteins differ in FRET-detected folding far inside the ribosome and in their exposure to ribosomal proteins, Cell, 116, 725-736.
Lu, J., and Deutsch, C. (2005) Folding zones inside the ribosomal exit tunnel, Nat. Struct. Mol. Biol., 12, 1123-1129.
Wilson, D. N., and Beckmann, R. (2011) The ribosomal tunnel as a functional environment for nascent polypeptide folding and translational stalling, Curr. Opin. Struct. Biol., 21, 274-282.
Tu, L., and Deutsch, C. (2017) Determinants of helix formation for a Kv1.3 transmembrane segment inside the ribosome exit tunnel, J. Mol. Biol., 429, 1722-1732.
Gilbert, R. J., Fucini, P., Connell, S., Fuller, S. D., Nierhaus, K. H., et al. (2004) Three-dimensional structures of translating ribosomes by Cryo-EM, Mol. Cell, 14, 57-66.
Kosolapov, A., and Deutsch, C. (2009) Tertiary interactions within the ribosomal exit tunnel, Nat. Struct. Mol. Biol., 16, 405-411.
Tu, L., Khanna, P., and Deutsch, C. (2014) Transmembrane segments form tertiary hairpins in the folding vestibule of the ribosome, J. Mol. Biol., 426, 185-198.
Holtkamp, W., Kokic, G., Jäger, M., Mittelstaet, J., Komar, A. A., and Rodnina, M. V. (2015) Cotranslational protein folding on the ribosome monitored in real time, Science, 350, 1104-1107.
Chen, X., Rajasekaran, N., Liu, K., and Kaiser, C. M. (2020) Synthesis runs counter to directional folding of a nascent protein domain, Nat. Commun., 11, 5096.
Komar, A. A. (2018) The Yin and Yang of codon usage, Hum. Mol. Genet., 25(R2), R77-R85.
Komar, A. A. (2007) SNPs, silent but not invisible, Science, 315, 466-467.
Tsai, C. J., Sauna, Z. E., Kimchi-Sarfaty, C., Ambudkar, S. V., Gottesman, M. M., and Nussinov, R. (2008) Synonymous mutations and ribosome stalling can lead to altered folding pathways and distinct minima, J. Mol. Biol., 383, 281-291.
Zhang, G., and Ignatova, Z. (2011) Folding at the birth of the nascent chain: coordinating translation with co-translational folding, Curr. Opin. Struct. Biol., 21, 25-31.
O’Brien, E. P., Ciryam, P., Vendruscolo, M., and Dobson, C. M. (2014) Understanding the influence of codon translation rates on cotranslational protein folding, Acc. Chem. Res., 47, 1536-1544.
Chaney, J. L., and Clark, P. L. (2015) Roles for synonymous codon usage in protein biogenesis, Annu. Rev. Biophys., 44, 143-166.
Jacobson, G. N., and Clark, P. L. (2016) Quality over quantity: optimizing co-translational protein folding with non-“optimal” synonymous codons, Curr. Opin. Struct. Biol., 38, 102-110.
Sharma, A. K., and O’Brien, E. P. (2018) Non-equilibrium coupling of protein structure and function to translation-elongation kinetics, Curr. Opin. Struct. Biol., 49, 94-103.
Samatova, E., Daberger, J., Liutkute, M., and Rodnina, M. V. (2021) Translational control by ribosome pausing in bacteria: how a non-uniform pace of translation affects protein production and folding, Front. Microbiol., 11, 619430.
Liu, Y., Yang, Q., and Zhao, F. (2021) Synonymous but not silent: the codon usage code for gene expression and protein folding, Annu. Rev. Biochem., 90, 375-401, https://doi.org/10.1146/annurev-biochem-071320-112701.
Pelham, H. R., and Jackson, R. J. (1976) An efficient mRNA-dependent translation system from reticulocyte lysates, Eur. J. Biochem., 67, 247-256.
Kolb, V. A., Makeyev, E. V., and Spirin, A. S. (1994) Folding of firefly luciferase during translation in a cell-free system, EMBO J., 13, 3631-3637.
Komar, A. A., Kommer, A., Krasheninnikov, I. A., and Spirin, A. S. (1993) Cotranslational heme binding to nascent globin chains, FEBS Lett., 326, 261-263.
Komar, A. A., Kommer, A., Krasheninnikov, I. A., and Spirin, A. S. (1997) Cotranslational folding of globin, J. Biol. Chem., 272, 10646-10651.
Purvis, I. J., Bettany, A. J., Santiago, T. C., Coggins, J. R., Duncan, K., et al. (1987) The efficiency of folding of some proteins is increased by controlled rates of translation in vivo. A hypothesis, J. Mol. Biol., 193, 413-417.
Krasheninnikov, I. A., Komar, A. A., and Adzhubeĭ, I. A. (1988) Role of the rare codon clusters in defining the boundaries of polypeptide chain regions with identical secondary structures in the process of co-translational folding of proteins [in Russian], Dokl. Akad. Nauk SSSR, 303, 995-999.
Krasheninnikov, I. A., Komar, A. A., and Adzhubeĭ, I. A. (1989) Frequency of using codons in mRNA and coding of the domain structure of proteins [in Russian], Dokl. Akad. Nauk. SSSR, 305, 1006-1012.
Krasheninnikov, I. A., Komar, A. A., and Adzhubeĭ, I. A. (1989) Role of the code redundancy determining cotranslational protein folding, Biokhimiia, 5, 187-200.
Heinemann, U., and Hahn, M. (1995) Circular permutation of polypeptide chains: implications for protein folding and stability, Prog. Biophys. Mol. Biol., 64, 121-143.
Protzel, A., and Morris, A. J. (1974) Gel chromatographic analysis of nascent globin chains. Evidence of nonuniform size distribution, J. Biol. Chem., 249, 4594-4600.
Chaney, W. G., and Morris, A. J. (1978) Nonuniform size distribution of nascent peptides: the role of messenger RNA, Arch. Biochem. Biophys., 191, 734-741.
Lizardi, P. M., Mahdavi, V., Shields, D., and Candelas, G. (1979) Discontinuous translation of silk fibroin in a reticulocyte cell-free system and in intact silk gland cells, Proc. Natl. Acad. Sci. USA, 76, 6211-6215.
Randall, L. L., Josefsson, L. G., and Hardy, S. J. (1980) Novel intermediates in the synthesis of maltose-binding protein in Escherichia coli, Eur. J. Biochem., 107, 375-379.
Abraham, A. K., and Pihl, A. (1980) Variable rate of polypeptide chain elongation in vitro. Effect of spermidine, Eur. J. Biochem., 106, 257-262.
Varenne, S., Knibiehler, M., Cavard, D., Morlon, J., and Lazdunski, C. (1982) Variable rate of polypeptide chain elongation for colicins A, E2 and E3, J. Mol. Biol., 159, 57-70.
Candelas, G., Candelas, T., Ortiz, A., and Rodríguez, O. (1983) Translational pauses during a spider fibroin synthesis, Biochem. Biophys. Res. Commun., 116, 1033-1038.
Varenne, S., Buc, J., Lloubes, R., and Lazdunski, C. (1984) Translation is a non-uniform process. Effect of tRNA availability on the rate of elongation of nascent polypeptide chains, J. Mol. Biol., 180, 549-576.
Wolin, S. L., and Walter, P. (1988) Ribosome pausing and stacking during translation of a eukaryotic mRNA, EMBO J., 7, 3559-3569.
Krasheninnikov, I. A., Komar, A. A., and Adzhubeĭ, I. A. (1991) Nonuniform size distribution of nascent globin peptides, evidence for pause localization sites, and a cotranslational protein-folding model, J. Protein. Chem., 10, 445-454.
Thanaraj, T. A., and Argos, P. (1996) Ribosome-mediated translational pause and protein domain organization, Protein Sci., 5, 1594-1612.
Thanaraj, T. A., and Argos, P. (1996) Protein secondary structural types are differentially coded on messenger RNA, Protein Sci., 5, 1973-1983.
Adzhubei, A. A., Adzhubei, I. A., Krasheninnikov, I. A., and Neidle, S. (1996) Non-random usage of “degenerate” codons is related to protein three-dimensional structure, FEBS Lett., 399, 78-82.
Oresic, M., and Shalloway, D. (1998) Specific correlations between relative synonymous codon usage and protein secondary structure, J. Mol. Biol., 281, 31-48.
Widmann, M., Clairo, M., Dippon, J., and Pleiss, J. (2008) Analysis of the distribution of functionally relevant rare codons, BMC Genomics, 9, 207.
Clarke, T. F. 4th, and Clark, P. L. (2008) Rare codons cluster, PLoS One, 3, e3412.
Chartier, M., Gaudreault, F., and Najmanovich, R. (2012) Large-scale analysis of conserved rare codon clusters suggests an involvement in co-translational molecular recognition events, Bioinformatics, 28, 1438-1445.
Ma, X. X., Wang, Y. N., Cao, X. A., Li, X. R., Liu, Y. S., et al. (2018) The effects of codon usage on the formation of secondary structures of nucleocapsid protein of peste des petits ruminants virus, Genes Genomics, 40, 905-912.
Newaz, K., Wright, G., Piland, J., Li, J., Clark, P. L., et al. (2020) Network analysis of synonymous codon usage, Bioinformatics, 36, 4876-4884.
McKown, R. L., Raab, R. W., Kachelries, P., Caldwell, S., and Laurie, G. W. (2013) Conserved regional 3′ grouping of rare codons in the coding sequence of ocular prosecretory mitogen lacritin, Invest. Ophthalmol. Vis. Sci., 54, 1979-1987.
Nissley, D. A., Carbery, A., Chonofsky, M., and Deane, C. M. (2021) Ribosome occupancy profiles are conserved between structurally and evolutionarily related yeast domains, Bioinformatics, https://doi.org/10.1093/bioinformatics/btab020.
Komar, A. A., and Jaenicke, R. (1995) Kinetics of translation of gamma B crystallin and its circularly permutated variant in an in vitro cell-free system: possible relations to codon distribution and protein folding, FEBS Lett., 376, 195-198.
Rudolph, R., Siebendritt, R., Nesslaŭer, G., Sharma, A. K., and Jaenicke, R. (1990) Folding of an all-beta protein: independent domain folding in gamma II-crystallin from calf eye lens, Proc. Natl. Acad. Sci. USA, 87, 4625-4629.
Komar, A. A., Lesnik, T., and Reiss, C. (1999) Synonymous codon substitutions affect ribosome traffic and protein folding during in vitro translation, FEBS Lett., 462, 387-391.
Kimchi-Sarfaty, C., Oh, J. M., Kim, I. W., Sauna, Z. E., Calcagno, A. M., et al. (2007) A “silent” polymorphism in the MDR1 gene changes substrate specificity, Science, 315, 525-528.
Zhang, G., Hubalewska, M., and Ignatova, Z. (2009) Transient ribosomal attenuation coordinates protein synthesis and co-translational folding, Nat. Struct. Mol. Biol., 16, 274-280.
Zhou, M., Guo, J., Cha, J., Chae, M., Chen, S., et al. (2013) Non-optimal codon usage affects expression, structure and function of clock protein FRQ, Nature, 495, 111-115.
Sander, I. M., Chaney, J. L., and Clark, P. L. (2014) Expanding Anfinsen’s principle: contributions of synonymous codon selection to rational protein design, J. Am. Chem. Soc., 136, 858-861.
Hu, S., Wang, M., Cai, G., and He, M. (2013) Genetic code-guided protein synthesis and folding in Escherichia coli, J. Biol. Chem., 288, 30855-30861.
Kim, S. J., Yoon, J. S., Shishido, H., Yang, Z., Rooney, L. A., et al. (2015) Protein folding. Translational tuning optimizes nascent protein folding in cells, Science, 348, 444-448.
Yu, C. H., Dang, Y., Zhou, Z., Wu, C., Zhao, F., et al. (2015) Codon usage influences the local rate of translation elongation to regulate co-translational protein folding, Mol. Cell, 59, 744-754.
Walsh, I. M., Bowman, M. A., Soto Santarriaga, I. F., Rodriguez, A., and Clark, P. L. (2020) Synonymous codon substitutions perturb cotranslational protein folding in vivo and impair cell fitness, Proc. Natl. Acad. Sci. USA, 117, 3528-3534.
Komar, A. A. (2007) Silent SNPs: impact on gene function and phenotype, Pharmacogenomics, 8, 1075-1080.
Sauna, Z. E., and Kimchi-Sarfaty, C. (2011) Understanding the contribution of synonymous mutations to human disease, Nat. Rev. Genet., 12, 683-691.
Hunt, R. C., Simhadri, V. L., Iandoli, M., Sauna, Z. E., and Kimchi-Sarfaty, C. (2014) Exposing synonymous mutations, Trends Genet., 30, 308-321.
Simhadri, V. L., Hamasaki-Katagiri, N., Lin, B. C., Hunt, R., Jha, S., et al. (2017) Single synonymous mutation in factor IX alters protein properties and underlies haemophilia B, J. Med. Genet., 54, 338-345.
Pechmann, S., Chartron, J. W., and Frydman, J. (2014) Local slowdown of translation by nonoptimal codons promotes nascent-chain recognition by SRP in vivo, Nat. Struct. Mol. Biol., 21, 1100-1105.
Buhr, F., Jha, S., Thommen, M., Mittelstaet, J., Kutz, F., et al. (2016) Synonymous codons direct cotranslational folding toward different protein conformations, Mol. Cell, 61, 341-351.
Kimchi-Sarfaty, C., Schiller, T., Hamasaki-Katagiri, N., Khan, M. A., Yanover, C., and Sauna, Z. E. (2013) Building better drugs: developing and regulating engineered therapeutic proteins, Trends Pharmacol. Sci., 34, 534-548.
Komar, A. A. (2016) The art of gene redesign and recombinant protein production: approaches and perspectives, Top. Med. Chem., 2, 1-17.
Alexaki, A., Hettiarachchi, G. K., Athey, J. C., Katneni, U. K., Simhadri, V., et al. (2019) Effects of codon optimization on coagulation factor IX translation and structure: implications for protein and gene therapies, Sci. Rep., 9, 15449.
Hunt, R., Hettiarachchi, G., Katneni, U., Hernandez, N., Holcomb, D., et al. (2019) A single synonymous variant (c.354G>A [p.P118P]) in ADAMTS13 confers enhanced specific activity, Int. J. Mol. Sci., 20, 5734.
Knobe, K. E., Sjorin, E., and Ljung, R. C. (2008) Why does the mutation G17736A/Val107Val (silent) in the F9 gene cause mild haemophilia B in five Swedish families?, Haemophilia, 14, 723-728.
Gartner, J. J., Parker, S. C., Prickett, T. D., Dutton-Regester, K., Stitzel, M. L., et al. (2013) Whole-genome sequencing identifies a recurrent functional synonymous mutation in melanoma, Proc. Natl. Acad. Sci. USA, 110, 13481-13486.
Hamasaki-Katagiri, N., Salari, R., Wu, A., Qi, Y., Schiller, T., et al. (2013) A gene-specific method for predicting hemophilia-causing point mutations, J. Mol. Biol., 425, 4023-4033.
Hamasaki-Katagiri, N., Lin, B. C., Simon, J., Hunt, R. C., Schiller, T., et al. (2017) The importance of mRNA structure in determining the pathogenicity of synonymous and non-synonymous mutations in haemophilia, Haemophilia, 23, e8-e17.
Katneni, U. K., Liss, A., Holcomb, D., Katagiri, N. H., Hunt, R., et al. (2019) Splicing dysregulation contributes to the pathogenicity of several F9 exonic point variants, Mol. Genet. Genomic Med., 7, e840.
Edwards, N. C., Hing, Z. A., Perry, A., Blaisdell, A., Kopelman, D. B., F et al. (2012) Characterization of coding synonymous and non-synonymous variants in ADAMTS13 using ex vivo and in silico approaches, PLoS One, 7, e38864.
Sauna, Z. E., Richards, S. M., Maillere, B., Jury, E. C., and Rosenberg, A. S. (2020) Editorial: immunogenicity of proteins used as therapeutics, Front. Immunol., 11, 614856.
Acknowledgments
This work would not be possible without the original contribution of Ivan Adzhubei and Igor A. Krasheninnikov, further support from Slava Kolb, Aigar Kommer, Valentin M. Stepanov, Lev P. Ovchinnikov, and Alexander S. Spirin, followed by collaborations with Rainer Jaenicke, Claude Reiss and more recently with Chava Kimchi-Sarfaty, Harald Schwalbe and Marina V. Rodnina.
I am indebted to all my teachers, colleagues, and collaborators for their extremely generous and inspiring discussions and invaluable contributions. I also apologize to those whose work or original publications could not be cited in this short review article.
Funding
In recent years, the work in my laboratory was financially supported by the Human Frontier Science Program Organization [HFSP grant #RGP0024/2010], the American Heart Association [AHA grant 13GRNT17070025], the National Institutes of Health [NIH grants HL121779; HL151392 and GM128981], the Center for Gene Regulation in Health and Disease (GRHD) at CSU, and the biotechnology company, DAPCEL, Inc.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
I am co-founder and Chief Scientific Officer of DAPCEL, Inc., a biotechnology company that develops innovative approaches for gene redesign for protein production in any desired host organism. This article does not contain any studies with human participants or animals performed by the author.
Rights and permissions
About this article
Cite this article
Komar, A.A. A Code Within a Code: How Codons Fine-Tune Protein Folding in the Cell. Biochemistry Moscow 86, 976–991 (2021). https://doi.org/10.1134/S0006297921080083
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0006297921080083