Abstract
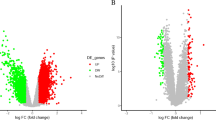
Ischemic stroke (IS) is a complex neurological disorder characterized by the sudden disruption of blood flow to the brain, leading to severe and often irreversible damage. Despite advances in stroke management, the underlying molecular mechanisms and key factors involved in the development and progression of IS remain elusive. In recent years, the integration of high-throughput data analysis techniques has emerged as a powerful approach to unraveling the molecular intricacies of complex diseases. In this study, we comprehensively analyzed gene expression, protein–protein interactions (PPI), and gene regulatory networks to identify IS-associated molecular factors. We utilized publicly available datasets and employed bioinformatics tools to analyze the data. Our analysis revealed many differentially expressed genes (DEGs) in IS, with a predominant down-regulation of genes. Gene ontology (GO) analysis highlighted the involvement of various biological processes, including transcriptional regulation, cell cycle, immune system processes, and cell differentiation. These findings underscore the complexity of stroke pathology, involving dysregulated gene expression and disrupted cellular processes. Constructing PPI networks enabled us to identify specific subnetworks associated with critical biological processes relevant to stroke, such as nucleosome assembly, protein translation, glycosylation, protein folding, and mRNA splicing. These subnetworks provide insights into the dysregulated molecular mechanisms contributing to stroke progression. Furthermore, we focused on identifying differentially expressed transcription factors (DE-TFs) within the gene regulatory network. Several up-regulated DE-TFs, including E2F1, MYB, GFI1B, and NUCKS1, were identified, suggesting their potential involvement in the dysregulation of gene expression in IS.




Similar content being viewed by others
Data Availability
Data used in this study is freely available and can be obtained from NCBI using above-mentioned GSE code “GSE22255.”
References
Peng L, et al. Microglia autophagy in ischemic stroke: a double-edged sword. Front Immunol. 2022;13:6825.
Zhao Y, et al. Neuronal injuries in cerebral infarction and ischemic stroke: from mechanisms to treatment. Int J Mol Med. 2022;49(2):1–9.
Campbell BC, et al. Ischaemic stroke. Nat Rev Dis Prim. 2019;5(1):70.
Bevan S, Markus HS. Genetics of common polygenic ischaemic stroke: current understanding and future challenges. Stroke Res Treat. 2011;2011.
Rutten-Jacobs LC et al. Genetic risk, incident stroke, and the benefits of adhering to a healthy lifestyle: cohort study of 306 473 UK Biobank participants. Bmj. 2018;363.
Adams HP Jr, et al. Classification of subtype of acute ischemic stroke. Definitions for use in a multicenter clinical trial. TOAST. Trial of Org 10172 in acute stroke treatment. Stroke. 1993;24(1):35–41.
Gasull T, Arboix A. Molecular mechanisms and pathophysiology of acute stroke: emphasis on biomarkers in the different stroke subtypes. 2022;9476 MDPI.
Zheng K, et al. Single-cell RNA-seq reveals the transcriptional landscape in ischemic stroke. J Cereb Blood Flow Metab. 2022;42(1):56–73.
Liu D-Z, et al. Brain and blood microRNA expression profiling of ischemic stroke, intracerebral hemorrhage, and kainate seizures. J Cereb Blood Flow Metab. 2010;30(1):92–101.
Xu B, et al. Analysis of expression profiles and bioinformatics suggests that plasma exosomal circular RNAs may be involved in ischemic stroke in the Chinese Han population. Metab Brain Dis. 2022;37(3):665–76.
Cho Y-E et al. Circulating immune cell landscape in patients who had mild ischaemic stroke. Stroke Vasc Neurol. 2022;7(4).
Ri MH et al. Regulatory mechanisms of natural compounds from traditional Chinese herbal medicines on the microglial response in ischemic stroke. Phytomedicine. 2023;154889.
He Q et al. Biological functions and regulatory mechanisms of hypoxia-inducible factor-1α in ischemic stroke. Front Immunol 2021;5377.
Yang K, et al. The mechanism of ferroptosis regulating oxidative stress in ischemic stroke and the regulation mechanism of natural pharmacological active components. Biomed Pharmacother. 2022;154:113611.
Chen L, et al. The protection by octreotide against experimental ischemic stroke: up-regulated transcription factor Nrf2, HO-1 and down-regulated NF-κB expression. Brain Res. 2012;1475:80–7.
Waseem A et al. Insight into the transcription factors regulating ischemic stroke and glioma in response to shared stimuli. in Seminars in Cancer Biology. 2023. Elsevier.
Mazur A, et al. Nrf2 as a therapeutic target in ischemic stroke. Expert Opin Ther Targets. 2021;25(3):163–6.
Patel S, et al. Pyruvate kinase M2 in chronic inflammations: a potpourri of crucial protein–protein interactions. Cell Biol Toxicol. 2021;37:653–78.
Szklarczyk D et al. The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2016;gkw937.
Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504.
Bastian M, Heymann S, Jacomy M. Gephi: an open source software for exploring and manipulating networks. in Proceedings of the international AAAI conference on web and social media. 2009.
Nepusz T, Yu H, Paccanaro A. Detecting overlapping protein complexes in protein-protein interaction networks. Nat Methods. 2012;9(5):471–2.
Lachmann A, et al. ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics. 2010;26(19):2438–44.
Huang DW et al. Extracting biological meaning from large gene lists with DAVID. Curr Protoc Bioinforma. 2009;27(1):13.11. 1–13.11. 13.
Sarkar S, et al. Cerebral ischemic stroke: cellular fate and therapeutic opportunities. Front Biosci-Landmark. 2019;24(3):415–30.
Jayaraj RL, et al. Neuroinflammation: friend and foe for ischemic stroke. J Neuroinflammation. 2019;16(1):1–24.
Rudilosso S, et al. The potential impact of neuroimaging and translational research on the clinical management of lacunar stroke. Int J Mol Sci. 2022;23(3):1497.
Regenhardt RW, et al. Advances in understanding the pathophysiology of lacunar stroke: a review. JAMA Neurol. 2018;75(10):1273–81.
Bierman-Chow S, et al. Clinical, imaging, and laboratory findings in patients with GATA2 deficiency presenting with early-onset ischemic stroke. Neurology. 2023;100(7):338–41.
Meadows C, Girotra T. An unusual manifestation of ischemic stroke in a patient with rare MonoMAC syndrome caused by N371K GATA2 mutation (2819). 2021;AAN Enterprises.
Sun J, et al. Krüppel-like factor 6 silencing prevents oxidative stress and neurological dysfunction following intracerebral hemorrhage via sirtuin 5/Nrf2/HO-1 axis. Front Aging Neurosci. 2021;13:646729.
Song L, et al. Li, Q. Q., Liu Q. Neuroprotective effects of SMADs in a rat model of cerebral ischemia/reperfusion. Neural Regen Res. 2015;10:438.
Wang Y-Z, et al. Association between PPARG genetic polymorphisms and ischemic stroke risk in a northern Chinese Han population: a case-control study. Neural Regen Res. 2019;14(11):1986.
Cheng F, et al. Relationship between PPAR-γ gene polymorphisms and ischemic stroke risk: a meta-analysis. Brain Behav. 2021;11(12):e2434.
Chen C, et al. CircRNA CTNNB1 (circCTNNB1) ameliorates cerebral ischemia/reperfusion injury by sponging miR-96-5p to up-regulate scavenger receptor class B type 1 (SRB1) expression. Bioengineered. 2022;13(4):10258–73.
Diaz-Cañestro C, et al. AP-1 (activated protein-1) transcription factor JunD regulates ischemia/reperfusion brain damage via IL-1β (interleukin-1β). Stroke. 2019;50(2):469–77.
Lian L, et al. Neuroinflammation in ischemic stroke: focus on microRNA-mediated polarization of microglia. Front Mol Neurosci. 2020;13:612439.
Ye J et al. Knockdown of ATF3 suppresses the progression of ischemic stroke through inhibiting ferroptosis. Front Mol Neurosci. 2023;15.
Author information
Authors and Affiliations
Contributions
MR and HF conceived the idea. MR conducted the experiment and prepared the results. HF supervised the procedure and verified the results and discussion. MR wrote the first draft, and HF edited the manuscript. MR and HF prepared the final draft.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Radak, M., Fallahi, H. Identification and prediction of molecular factors associated with ischemic stroke: an integrative analysis of DEGs, TFs, and PPI networks. In vitro models 2, 307–315 (2023). https://doi.org/10.1007/s44164-023-00063-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s44164-023-00063-y