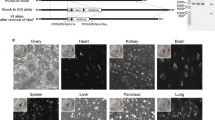
Abstract
Gene expression during spermatogenesis undergoes significant changes due to a demanding sequence of mitosis, meiosis, and differentiation. We investigated the contribution of H3 histone modifications to gene regulation in the round spermatids. Round spermatids were purified from rat testes using centrifugal elutriation and Percoll density-gradient centrifugation. After enzymatic chromatin shearing, immuno-precipitation using antibodies against histone marks H3k4me3 and H3K9me3 was undertaken. The immunoprecipitated DNA fragments were subjected to massive parallel sequencing. Gene expression in round spermatids and sperm was analyzed by transcriptome sequencing using next-generation sequencing methods. ChIP-seq analysis showed significant peak enrichment in H3K4me3 marks in active chromatin regions and H3K9me3 peak enrichment in repressive regions. We found 53 genes which showed overlapping peak enrichment in both H3K4me3 and H3K9me3 marks. Some of the top H3K4me3-enriched genes were involved in sperm tail formation (Odf1, Odf3, Odf4, Oaz3, Ccdc42, Ccdc63, and Ccdc181), chromatin condensation (Dync1h1, Dynll1, and Kdm3a), and sperm functions such as acrosome reaction (Acrbp and Fabp9), energy generation (Gapdhs), and signaling for motility (Tssk1b, Tssk2, and Tssk4). Transcriptome sequencing in round spermatids found 64% transcripts of the H3K4me3-enriched genes at high levels and of about 25% of H3K9me3-enriched genes at very low levels. Transcriptome sequencing in sperm found that more than 99% of the ChIP-seq corresponding transcripts were also present in sperm. H3K4me3 enrichment in the round spermatids correlates significantly with gene expression and H3K9me3 correlates with gene silencing that contribute to sperm differentiation and setting the RNA payloads of sperm.







Similar content being viewed by others
Data Availability
All representative data are provided within the manuscript and also in the supporting information. Sequencing data can be accessed by GEO accession number GSE151608.
Code Availability
Not applicable.
References
Pandey A, Yadav SK, Vishvkarma R, Singh B, Maikhuri JP, Rajender S, Gupta, G. The dynamics of gene expression during and post meiosis sets the sperm agenda. Mol Reprod Dev 2019;
Kleene KCA. possible meiotic function of the peculiar patterns of gene expression in mammalian spermatogenic cells. Mech Dev. 2001;106:3–23.
Gatewood JM, Cook GR, Balhorn R, Schmid C, Bradbury EM. Isolation of four core histones from human sperm chromatin representing a minor subset of somatic histones. J Biol Chem. 1990;265:20662–6.
Wang T, Gao H, Li W, Liu C. Essential role of histone replacement and modifications in male fertility. Front Genet. 2019;8(10):962.
Bao J, Bedford MT. Epigenetic regulation of the histone-to-protamine transition during spermiogenesis. Reproduction (Cambridge, England). 2016;151(5):R55.
Yuen BT, Bush KM, Barrilleaux BL, Cotterman R, Knoepfler PS. Histone H3. 3 regulates dynamic chromatin states during spermatogenesis. Development. 2014;141(18):3483–94.
Shilatifard A. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem. 2012;81:65–95.
Lauberth SM, Nakayama T, Wu X, Ferris AL, Tang Z, Hughes SH, Roeder RG. H3K4me3 interactions with TAF3 regulate preinitiation complex assembly and selective gene activation. Cell. 2013;152:1021–36.
Flanagan JF, Mi LZ, Chruszcz M, Cymborowski M, Clines KL, Kim Y, Minor W, Rastinejad F, Khorasanizadeh S. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature. 2005;438:1181–5.
Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–5.
Nishioka K, Chuikov S, Sarma K, Erdjument-Bromage H, Allis CD, Tempst P, Reinberg D. Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes Dev. 2002;16:479–89.
Schneider R, Bannister AJ, Weise C, Kouzarides T. Direct binding of INHAT to H3 tails disrupted by modifications. J Biol Chem. 2004;279:23859–62.
Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–37.
Lehnertz B, Ueda Y, Derijck AA, Braunschweig U, Perez-Burgos L, Kubicek S, Chen T, Li E, Jenuwein T, Peters AH. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol. 2003;13:1192–200.
Otani J, Nankumo T, Arita K, Inamoto S, Ariyoshi M, Shirakawa M. Structural basis for recognition of H3K4 methylation status by the DNA methyltransferase 3A ATRX–DNMT3–DNMT3L domain. EMBO Rep. 2009;10(11):1235–41.
Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460:473–8.
Yadav SK, Pandey A, Kumar L, Devi A, Kushwaha B, Vishvkarma R, Maikhuri JP, Rajender S, Gupta G. The thermo-sensitive gene expression signatures of spermatogenesis. Reprod Biol Endocrinol. 2018;16:56.
Mercier E, Droit A, Li L, Robertson G, Zhang X, Gottardo R. An integrated pipeline for the genome-wide analysis of transcription factor binding sites from ChIP-Seq. PLoS One 2011;6:e16432
Yu G, Wang LG, He QY. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015;31:2382–3.
Yu G, He QY. ReactomePA: an R/Bioconductor package for reactome pathway analysis and visualization. Mol Biosyst. 2016;12:477–9.
Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–7.
Hou CC, Yang WX. New insights to the ubiquitin-proteasome pathway (UPP) mechanism during spermatogenesis. Mol Biol Rep. 2013;40:3213–30.
Escalier D, Bai XY, Silvius D, Xu PX, Xu X. Spermatid nuclear and sperm periaxonemal anomalies in the mouse Ube2b null mutant. Mol Reprod Dev. 2003;65:298–308.
Suryavathi V, Khattri A, Gopal K, Rani DS, Panneerdoss S, Gupta NJ, Chakravarty B, Deenadayal M, Singh L, Thangaraj K. Novel variants in UBE2B gene and idiopathic male infertility. J Androl. 2008;29:564–71.
Yatsenko AN, Georgiadis AP, Murthy LJ, Lamb DJ, Matzuk MM. UBE2B mRNA alterations are associated with severe oligozoospermia in infertile men. Mol Hum Reprod. 2013;19:388–94.
Nakamura N. Ubiquitination regulates the morphogenesis and function of sperm organelles. Cells. 2013;2:732–50.
Peters AH, O’Carroll D, Scherthan H, Mechtler K, Sauer S, Schofer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, Opravil S, Doyle M, Sibilia M, Jenuwein T. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–37.
O’Carroll D, Scherthan H, Peters AH, Opravil S, Haynes AR, Laible G, Rea S, Schmid M, Lebersorger A, Jerratsch M, Sattler L, Mattei MG, Denny P, Brown SD, Schweizer D, Jenuwein T. Isolation and characterization of Suv39h2, a second histone H3 methyltransferase gene that displays testis-specific expression. Mol Cell Biol. 2000;20:9423–33.
Soufi A, Donahue G, Zaret KS. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell. 2012;151:994–1004.
Lesch BJ, Silber SJ, McCarrey JR, Page DC. Parallel evolution of male germline epigenetic poising and somatic development in animals. Nat Genet. 2016;48(8):888–94. Erratum in: Nat Genet. 2016;28;48(10):1296.
Wang X, Kang JY, Wei L, Yang X, Sun H, Yang S, Lu L, Yan M, Bai M, Chen Y, Long J, Li N, Li D, Huang J, Lei M, Shao Z, Yuan W, Zuo E, Lu K, Liu MF, Li J. PHF7 is a novel histone H2A E3 ligase prior to histone-to-protamine exchange during spermiogenesis. Development. 2019 10;146(13):dev175547. Erratum in: Development. 2020;147(8): PMID: 31189663.
Maezawa S, Hasegawa K, Yukawa M, Kubo N, Sakashita A, Alavattam KG, Sin HS, Kartashov AV, Sasaki H, Barski A, Namekawa SH. Polycomb protein SCML2 facilitates H3K27me3 to establish bivalent domains in the male germline. Proc Natl Acad Sci U S A. 2018;115(19):4957–62.
Erkek S, Hisano M, Liang CY, Gill M, Murr R, Dieker J, Schübeler D, van der Vlag J, Stadler MB, Peters AH. Molecular determinants of nucleosome retention at CpG-rich sequences in mouse spermatozoa. Nat Struct Mol Biol. 2013;20(7):868–75. Erratum in: Nat Struct Mol Biol. 2013 ;20(10):1236.
Liu Y, Zhang Y, Yin J, Gao Y, Li Y, Bai D, He W, Li X, Zhang P, Li R, Zhang L, Jia Y, Zhang Y, Lin J, Zheng Y, Wang H, Gao S, Zeng W, Liu W. Distinct H3K9me3 and DNA methylation modifications during mouse spermatogenesis. J Biol Chem. 2019;294(49):18714–25.
Bryant JM, Donahue G, Wang X, Meyer-Ficca M, Luense LJ, Weller AH, Bartolomei MS, Blobel GA, Meyer RG, Garcia BA, Berger SL. Characterization of BRD4 during mammalian postmeiotic sperm development. Mol Cell Biol. 2015;35(8):1433–48.
Teperek M, Simeone A, Gaggioli V, Miyamoto K, Allen GE, Erkek S, Kwon T, Marcotte EM, Zegerman P, Bradshaw CR, Peters AH, Gurdon JB, Jullien J. Sperm is epigenetically programmed to regulate gene transcription in embryos. Genome Res. 2016;26:1034–46.
Oikawa M, Simeone A, Hormanseder E, Teperek M, Gaggioli V, O’Doherty A, Falk E, Sporniak M, D’Santos C, Franklin VN, Kishore K. Epigenetic homogeneity in histone methylation underlies sperm programming for embryonic transcription. Nat Commun. 2020;11(1):1–6.
Harbuz R, Zouari R, Pierre V, Ben Khelifa M, Kharouf M, Coutton C, Merdassi G, Abada F. Escoffier J, Nikas Y, Vialard F, Koscinski I, Triki C, et al. A recurrent deletion of DPY19L2 causes infertility in man by blocking sperm head elongation and acrosome formation. Am J Hum Genet 2011;88:351–361
Doran J, Walters C, Kyle V, Wooding P, Hammett-Burke R, Colledge WH. Mfsd14a (Hiat1) gene disruption causes globozoospermia and infertility in male mice. Reproduction. 2016;152:91–9.
Takasaki N, Tachibana K, Ogasawara S, Matsuzaki H, Hagiuda J, Ishikawa H, Mochida K, Inoue K, Ogonuki N, Ogura A, Noce T, Ito C, Toshimori K, Narimatsu H. A heterozygous mutation of GALNTL5 affects male infertility with impairment of sperm motility. Proc Natl Acad Sci U S A. 2014;111:1120–5.
Escoffier J, Lee HC, Yassine S, Zouari R, Martinez G, Karaouzene T, Coutton C, Kherraf ZE, Halouani L, Triki C, Nef S, Thierry-Mieg N, Savinov SN, Fissore R, Ray PF, Arnoult C. Homozygous mutation of PLCZ1 leads to defective human oocyte activation and infertility that is not rescued by the WW-binding protein PAWP. Hum Mol Genet. 2016;25:878–91.
Zhu F, Liu C, Wang F, Yang X, Zhang J, Wu H, Zhang Z, He X, Zhang Z, Zhou P, Wei Z, Shang Y, Wang L, Zhang R, Ouyang YC, Sun QY, Cao Y, Li W. Mutations in PMFBP1 cause acephalic spermatozoa syndrome. Am J Hum Genet. 2018;103:188–99.
Okutman O, Muller J, Skory V, Garnier JM, Gaucherot A, Baert Y, Lamour V, Serdarogullari M, Gultomruk M, Ropke A, Kliesch S, Herbepin V, et al. A no-stop mutation in MAGEB4 is a possible cause of rare X-linked azoospermia and oligozoospermia in a consanguineous Turkish family. J Assist Reprod Genet. 2017;34:683–94.
Jimenez A, Zu W, Rawe VY, Pelto-Huikko M, Flickinger CJ, Sutovsky P, Gustafsson JA, Oko R, Miranda-Vizuete A. 2Spermatocyte/spermatid-specific thioredoxin-3, a novel Golgi apparatus-associated thioredoxin, is a specific marker of aberrant spermatogenesis. J Biol Chem. 2004;279:34971–82.
Esfahani MH, Abbasi H, Mirhosseini Z, Ghasemi N, Razavi S, Tavalaee M, Tanhaei S, Deemeh MR, Ghaedi K, Zamansoltani F, Rajaei F. Can altered expression of hspa2 in varicocele patients lead to abnormal spermatogenesis? International Journal of Fertility and Sterility. 2010;4(3):104–13.
Motiei M, Tavalaee M, Rabiei F, Hajihosseini R, Nasr-Esfahani MH. Evaluation of HSPA 2 in fertile and infertile individuals. Andrologia. 2013;45(1):66–72.
Funding
The authors received funding from the Council of Scientific and Industrial Research (CSIR) under the network scheme of projects (BSC0101). Poonam Mehta received a graduate fellowship from the University Grants Commission (460/CSIR-UGC NET DEC.2017).
Author information
Authors and Affiliations
Contributions
SS, RS and GG have conceived the idea. SS, SY and RS have performed the experimental procedures. SS, PM and RS have analyzed the data. SS, PM and RS wrote the manuscript.
Corresponding author
Ethics declarations
Ethics Approval
The study was approved by the institutional ethics committee of CSIR-CDRI (IAEC/2014/49/Renew03(135/16)) and the study was performed in accordance with the ethical standards as mentioned in the 1964 Declaration of Helsinki and its later amendments.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Competing Interests
The authors declare no competing interests.
Additional information
Council of Scientific and Industrial Research,India,BSC0101,Singh Rajender
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sarkar, S., Yadav, S., Mehta, P. et al. Histone Methylation Regulates Gene Expression in the Round Spermatids to Set the RNA Payloads of Sperm. Reprod. Sci. 29, 857–882 (2022). https://doi.org/10.1007/s43032-021-00837-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s43032-021-00837-3