Abstract
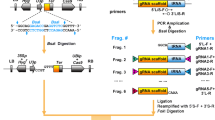
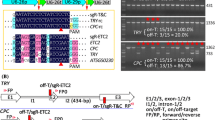
The CRISPR/Cas9 genome-editing system has emerged as a popular powerful tool for biological research. However, the process of selecting efficiently edited Cas9-free plants is usually laborious and time consuming. Here, we demonstrated P2A to be the most efficient self-cleaving peptide for fusing Cas9 and GFP in Arabidopsis and then used Cas9-P2A-GFP to develop a novel CRISPR/Cas9 system. Additionally, a pair of isocaudomer restriction enzymes were selected to conveniently assemble multiple sgRNAs. In this system, the GFP fluorescence intensity in T1 transgenic plants indicates the expression level of the Cas9 protein, which correlates well with the editing efficiency. Furthermore, Cas9-free plants can be easily selected by examining GFP fluorescence in T2 transgenic plants. The efficient knockout of BRI1, BZR1 and BES1 demonstrated the robustness of our new system. Thus, we designed a novel CRISPR/Cas9 system that can generate Cas9-free multiplex mutants efficiently in Arabidopsis and possibly in other plant species.



Similar content being viewed by others
References
Buren S, Ortega-Villasante C, Otvos K, Samuelsson G, Bako L, Villarejo A (2012) Use of the foot-and-mouth disease virus 2A peptide co-expression system to study intracellular protein trafficking in Arabidopsis. PLoS One 7:e51973
Cermak T, Curtin SJ, Gil-Humanes J, Cegan R, Kono TJY, Konecna E, Belanto JJ, Starker CG, Mathre JW, Greenstein RL, Voytas DF (2017) A multipurpose toolkit to enable advanced genome engineering in plants. Plant Cell 29:1196–1217
de Felipe P, Luke GA, Brown JD, Ryan MD (2010) Inhibition of 2A-mediated ‘cleavage’ of certain artificial polyproteins bearing N-terminal signal sequences. Biotechnol J 5:213–223
Donnelly MLL, Luke G, Mehrotra A, Li XJ, Hughes LE, Gani D, Ryan MD (2001) Analysis of the aphthovirus 2A/2B polyprotein ‘cleavage’ mechanism indicates not a proteolytic reaction, but a novel translational effect: a putative ribosomal ‘skip’. J Gen Virol 82:1013–1025
Feng ZY, Zhang BT, Ding WN, Liu XD, Yang DL, Wei PL, Cao FQ, Zhu SH, Zhang F, Mao YF, Zhu JK (2013) Efficient genome editing in plants using a CRISPR/Cas system. Cell Res 23:1229–1232
Feng ZY, Mao YF, Xu NF, Zhang BT, Wei PL, Yang DL, Wang Z, Zhang ZJ, Zheng R, Yang L, Zeng L, Liu XD, Zhu JK (2014) Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc Natl Acad Sci USA 111:4632–4637
Gao XH, Chen JL, Dai XH, Zhang D, Zhao YD (2016) An effective strategy for reliably isolating heritable and Cas9-free Arabidopsis mutants generated by CRISPR/Cas9-mediated genome editing. Plant Physiol 171:1794–1800
Horvath P, Barrangou R (2010) CRISPR/Cas, the immune system of bacteria and archaea. Science 327:167–170
Kim JH, Lee SR, Li LH, Park HJ, Park JH, Lee KY, Kim MK, Shin BA, Choi SY (2011) High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS One 6:e18556
Li J, Chory J (1997) A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90:929–938
Li JF, Norville JE, Aach J, McCormack M, Zhang DD, Bush J, Church GM, Sheen J (2013) Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotechnol 31:688–691
Ma XL, Zhang QY, Zhu QL, Liu W, Chen Y, Qiu R, Wang B, Yang ZF, Li HY, Lin YR, Xie YY, Shen RX, Chen SF, Wang Z, Guo JX, Chen LT, Zhao XC, Dong Z, Liu YG (2015) A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant 8:1274–1284
Mao YF, Zhang H, Xu NF, Zhang BT, Gou F, Zhu JK (2013) Application of the CRISPRCas system for efficient genome engineering in plants. Mol Plant 6:2008–2011
Mao YF, Zhang ZJ, Feng ZY, Wei PL, Zhang H, Botella JR, Zhu JK (2016) Development of germ-line-specific CRISPR-Cas9 systems to improve the production of heritable gene modifications in Arabidopsis. Plant Biotechnol J 14:519–532
Mao YF, Botella JR, Liu YG, Zhu JK (2019) Gene editing in plants: progress and challenges. Natl Sci Rev 6:421–437
Petersen BL, Moller SR, Mravec J, Jorgensen B, Christensen M, Liu Y, Wandall HH, Bennett EP, Yang Z (2019) Improved CRISPR/Cas9 gene editing by fluorescence activated cell sorting of green fluorescence protein tagged protoplasts. BMC Biotechnol 19:36
Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, Dahlman JE, Parnas O, Eisenhaure TM, Jovanovic M, Graham DB, Jhunjhunwala S, Heidenreich M, Xavier RJ, Langer R, Anderson DG, Hacohen N, Regev A, Feng G, Sharp PA, Zhang F (2014) CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159:440–455
Sun N, Wang JJ, Gao ZX, Dong J, He H, Terzaghi W, Wei N, Deng XW, Chen HD (2016) Arabidopsis SAURs are critical for differential light regulation of the development of various organs. Proc Natl Acad Sci USA 113:6071–6076
Wang C, Shen L, Fu YP, Yan CJ, Wang KJ (2015a) A simple CRISPR/Cas9 system for multiplex genome editing in rice. J Genet Genom 42:703–706
Wang ZP, Xing HL, Dong L, Zhang HY, Han CY, Wang XC, Chen QJ (2015b) Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol 16:144
Wang M, Lu Y, Botella JR, Mao Y, Hua K, Zhu JK (2017) Gene targeting by homology-directed repair in rice using a geminivirus-based CRISPR/Cas9 system. Mol Plant 10:1007–1010
Yan LH, Wei SW, Wu YR, Hu RL, Li HJ, Yang WC, Xie Q (2015) High-efficiency genome editing in Arabidopsis using YAO promoter-driven CRISPR/Cas9 system. Mol Plant 8:1820–1823
Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2:1565–1572
Yu H, Zhao YD (2019) Fluorescence marker-assisted isolation of Cas9-free and CRISPR-edited Arabidopsis plants. Methods Mol Biol 1917:147–154
Zhang ZJ, Mao YF, Ha S, Liu WS, Botella JR, Zhu JK (2016) A multiplex CRISPR/Cas9 platform for fast and efficient editing of multiple genes in Arabidopsis. Plant Cell Rep 35:1519–1533
Acknowledgements
We thank Prof. Xing Wang Deng for lab resources, Prof. Jian-Kang Zhu for providing the p35S-Cas9-SK and pAtU6-26-SK plasmids, and Prof. Qi-Jun Chen for providing the pHEE401E vector. This study was supported by the National Key R&D Program of China (2018YFE0204700), the National Natural Science Foundation of China (31621001), the Peking-Tsinghua Center for Life Sciences, and the State Key Laboratory of Protein and Plant Gene Research.
Author information
Authors and Affiliations
Contributions
JW and HC designed the study. JW performed the experiments. JW and HC analyzed the data. JW and HC wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, J., Chen, H. A novel CRISPR/Cas9 system for efficiently generating Cas9-free multiplex mutants in Arabidopsis. aBIOTECH 1, 6–14 (2020). https://doi.org/10.1007/s42994-019-00011-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42994-019-00011-z