Abstract
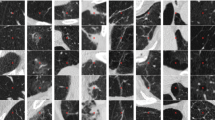
Interstitial lung disease (ILD) encompasses a spectrum of more than 200 fatal lung disorders affecting the interstitium, contributing to substantial mortality rates. The intricate process of diagnosing ILDs is compounded by their diverse symptomatology and resemblance to other pulmonary conditions. High-resolution computed tomography (HRCT) assumes the role of the primary diagnostic tool for ILD, playing a pivotal role in the medical landscape. In response, this study introduces a computational framework powered by artificial intelligence (AI) to support medical professionals in the identification and classification of ILD from HRCT images. Our dataset comprises 3045 HRCT images sourced from distinct patient cases. The proposed framework presents a novel approach to predicting ILD categories using a two-tier ensemble strategy that integrates outcomes from convolutional neural networks (CNNs), transfer learning, and machine learning (ML) models. This approach outperforms existing methods when evaluated on previously unseen data. Initially, ML models, including Logistic Regression, BayesNet, Stochastic Gradient Descent (SGD), RandomForest, and J48, are deployed to detect ILD based on statistical measures derived from HRCT images. Notably, the J48 model achieves a notable accuracy of 93.08%, with the diagnostic significance of diagonal-wise standard deviation emphasized through feature analysis. Further refinement is achieved through the application of Marker-controlled Watershed Transformation Segmentation and Morphological Masking techniques to HRCT images, elevating accuracy to 95.73% with the J48 model. The computational framework also embraces deep learning techniques, introducing three innovative CNN models that achieve test accuracies of 94.08%, 92.04%, and 93.72%. Additionally, we evaluate five full-training and transfer learning models (InceptionV3, VGG16, MobileNetV2, VGG19, and ResNet50), with the InceptionV3 model achieving peak accuracy at 78.41% for full training and 92.48% for transfer learning. In the concluding phase, a soft-voting ensemble mechanism amplifies training outcomes, yielding ensemble test accuracies of 76.56% for full-training models and 92.81% for transfer learning models. Notably, the ensemble comprising the three newly introduced CNN models attains the pinnacle of test accuracy at 97.42%. This research is poised to drive advancements in ILD diagnosis, presenting a resilient computational framework that enhances accuracy and ultimately betters patient outcomes within the medical domain.







Similar content being viewed by others
Data Availability
The data that support the findings of this study are available from St. John's Medical College, Bengaluru, India but restrictions apply to the availability of these data, which were used under licence for the current study, and so are not publicly available. Data are, however, available from the authors upon reasonable request and with permission of St. John's Medical College, Bengaluru, India.
References
Hodnett PA, Naidich DP. Fibrosing interstitial lung disease: a practical high-resolution computed tomography-based approach to diagnosis and management and a review of the literature. Am J Respir Crit Care Med. 2013;188(2):141–9. https://doi.org/10.1164/RCCM.201208-1544CI/SUPPL_FILE/DISCLOSURES.PDF.
Wells AU. The revised ATS/ERS/JRS/ALAT diagnostic criteria for idiopathic pulmonary fibrosis (IPF)—practical implications. Respir Res. 2023;14(Suppl 1):1–6. https://doi.org/10.1186/1465-9921-14-S1-S2/TABLES/4.
Fernández Pérez ER, et al. Incidence, prevalence, and clinical course of idiopathic pulmonary fibrosis: a population-based study. Chest. 2010;137(1):129–37. https://doi.org/10.1378/CHEST.09-1002.
Collard HR, King TE, Bartelson BB, Vourlekis JS, Schwarz MI, Brown KK. Changes in clinical and physiologic variables predict survival in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2012;168(5):538–42. https://doi.org/10.1164/RCCM.200211-1311OC.
Flaherty KR, et al. Idiopathic Interstitial Pneumonia. Am J Respir Crit Care Med. 2012;170(8):904–10. https://doi.org/10.1164/RCCM.200402-147OC.
Xu R, Hirano Y, Tachibana R, Kido S. Classification of diffuse lung disease patterns on high-resolution computed tomography by a bag of words approach. Lect Notes Comput Sci. 2011;6893(3):183–90. https://doi.org/10.1007/978-3-642-23626-6_23/COVER.
Gangeh MJ, Sørensen L, Shaker SB, Kamel MS, De Bruijne M, Loog M. A texton-based approach for the classification of lung parenchyma in CT images. Lect Notes Comput Sci. 2010;6363(3):595–602. https://doi.org/10.1007/978-3-642-15711-0_74/COVER.
Pradeep IK, Jaya Bhaskar M, Satyanarayana B. Data science and deep learning applications in the e-commerce industry: a survey. Indian J Comput Sci Eng. 2020;11(5):497–509.
Sivachandiran S, Jagan Mohan K, Mohammed Nazer G. Intelligent deep learning enabled crowd detection and classification model in real time surveillance videos.
Bejnordi BE, et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA. 2017;318(22):2199–210. https://doi.org/10.1001/JAMA.2017.14585.
Mcadams HP, Samei E, Dobbins J, Tourassi GD, Ravin CE. Recent advances in chest radiography. Radiology. 2006. https://doi.org/10.1148/radiol.2413051535.
Cicero M, et al. Training and validating a deep convolutional neural network for computer-aided detection and classification of abnormalities on frontal chest radiographs. Investig Radiol. 2017;52(5):281–7. https://doi.org/10.1097/RLI.0000000000000341.
Lakhani P, Sundaram B. Deep learning at chest radiography: automated classification of pulmonary tuberculosis by using convolutional neural networks. Radiology. 2017;284(2):574–82. https://doi.org/10.1148/RADIOL.2017162326.
Gonzalez G, et al. Disease staging and prognosis in smokers using deep learning in chest computed tomography. Am J Respir Crit Care Med. 2018;197(2):193–203. https://doi.org/10.1164/RCCM.201705-0860OC/SUPPL_FILE/DISCLOSURES.PDF.
Heitmann KR, Kauczor HU, Mildenberger P, Uthmann T, Perl J, Thelen M. Automatic detection of ground glass opacities on lung HRCT using multiple neural networks. Eur Radiol. 2014;7(9):1463–72. https://doi.org/10.1007/S003300050318.
Uppaluri R, Hoffman EA, Sonka M, Hartley PG, Hunninghake GW, McLennan G. Computer recognition of regional lung disease patterns. Am J Respir Crit Care Med. 2012;160(2):648–54. https://doi.org/10.1164/AJRCCM.160.2.9804094.
Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. Commun ACM. 2017;60(6):84–90. https://doi.org/10.1145/3065386.
van Tulder G, de Bruijne M. Learning features for tissue classification with the classification restricted Boltzmann machine. Lect Notes Comput Sci. 2014;8848:47–58. https://doi.org/10.1007/978-3-319-13972-2_5/COVER.
Li Q, Cai W, Wang X, Zhou Y, Feng DD, Chen M (2014) Medical image classification with convolutional neural network. In: 2014 13th Int. Conf. Control Autom. Robot. Vision, ICARCV 2014; 2014. p. 844–8. https://doi.org/10.1109/ICARCV.2014.7064414.
Gao M, et al. Holistic classification of CT attenuation patterns for interstitial lung diseases via deep convolutional neural networks. Comput Methods Biomech Biomed Eng. 2016;6(1):1–6. https://doi.org/10.1080/21681163.2015.1124249.
Anthimopoulos M, Christodoulidis S, Ebner L, Christe A, Mougiakakou S. Lung pattern classification for interstitial lung diseases using a deep convolutional neural network. IEEE Trans Med Imaging. 2016;35(5):1207–16. https://doi.org/10.1109/TMI.2016.2535865.
Christodoulidis S, Anthimopoulos M, Ebner L, Christe A, Mougiakakou S. Multisource transfer learning with convolutional neural networks for lung pattern analysis. IEEE J Biomed Health Inform. 2017;21(1):76–84. https://doi.org/10.1109/JBHI.2016.2636929.
Kim GB, et al. Comparison of shallow and deep learning methods on classifying the regional pattern of diffuse lung disease. J Digit Imaging. 2018;31(4):415–24. https://doi.org/10.1007/S10278-017-0028-9/FIGURES/5.
Wang Z, et al. Optimal threshold in CT quantification of emphysema. Eur Radiol. 2012;23(4):975–84. https://doi.org/10.1007/S00330-012-2683-Z.
Bae HJ, et al. A Perlin noise-based augmentation strategy for deep learning with small data samples of HRCT images. Sci Rep. 2018;8(1):1–7. https://doi.org/10.1038/s41598-018-36047-2.
Walsh SLF, Calandriello L, Silva M, Sverzellati N. Deep learning for classifying fibrotic lung disease on high-resolution computed tomography: a case-cohort study. Lancet Respir Med. 2018;6(11):837–45. https://doi.org/10.1016/S2213-2600(18)30286-8.
Sharif Razavian A, Azizpour H, Sullivan J, Carlsson S. CNN features off-the-shelf: an astounding baseline for recognition; 2014. p. 806–13.
Zheng L, Zhao Y, Wang S, Wang J, Tian Q. Good practice in CNN feature transfer (2016). https://doi.org/10.48550/arxiv.1604.00133.
Yosinski J, Clune J, Bengio Y, Lipson H. How transferable are features in deep neural networks? Adv Neural Inf Process Syst. 2014;27:3320–8.
Cheplygina V, Pena IP, Pedersen JH, Lynch DA, Sorensen L, De Bruijne M. Transfer learning for multicenter classification of chronic obstructive pulmonary disease. IEEE J Biomed Health Inform. 2018;22(5):1486–96. https://doi.org/10.1109/JBHI.2017.2769800.
Wei X, Chen J, Cai C. Using deep convolutional neural networks and transfer learning for mammography mass lesion classification. J Comput Theor Nanosci. 2017;14(8):3802–6. https://doi.org/10.1166/JCTN.2017.6676.
Yap MH, et al. Automated breast ultrasound lesions detection using convolutional neural networks. IEEE J Biomed Health Inform. 2018;22(4):1218–26. https://doi.org/10.1109/JBHI.2017.2731873.
Lu Y, Chen L, Saidi A. Optimal transport for deep joint transfer learning (2017). https://doi.org/10.48550/arxiv.1709.02995.
Samala RK, Chan HP, Hadjiiski L, Helvie MA, Richter CD, Cha KH. Breast cancer diagnosis in digital breast tomosynthesis: effects of training sample size on multi-stage transfer learning using deep neural nets. IEEE Trans Med Imaging. 2019;38(3):686–96. https://doi.org/10.1109/TMI.2018.2870343.
Suzuki A, Sakanashi H, Kido S, Shouno H. Feature representation analysis of deep convolutional neural network using two-stage feature transfer—an application for diffuse lung disease classification (2018). https://doi.org/10.48550/arxiv.1810.06282.
Raju AHBN, Augustine P. Identification of interstitial lung diseases using deep learning. Int J Electr Comput Eng. 2020;10(6):6283–91. https://doi.org/10.11591/ijece.v10i6.pp6283-6291.
Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z. Rethinking the inception architecture for computer vision; 2016. p. 2818–26.
Lin C, et al. Transfer learning based traffic sign recognition using inception-v3 model. Period Polytech Transp Eng. 2019;47(3):242–50.
Chollet F. Xception: deep learning with depthwise separable convolutions; 2017. p. 1251–8.
Dong K, et al. MobileNetV2 model for image classification. In: 2020 2nd International conference on information technology and computer application (ITCA). IEEE; 2020.
Han S, Mao H, Dally WJ. Deep compression: compressing deep neural networks with pruning, trained quantization and Huffman coding. In: 4th Int. Conf. Learn. Represent. ICLR 2016—Conf. Track Proc; 2015. https://doi.org/10.48550/arxiv.1510.00149.
Hridayami P, Ketut Gede Darma Putra I, Wibawa KS. Fish species recognition using VGG16 deep convolutional neural network. J Comput Sci Eng. 2019;13(3):124–30.
Understanding the VGG19 Architecture. https://iq.opengenus.org/vgg19-architecture/ (Accessed 8 Dec 2022).
Khan MSM, et al. Cataract detection using convolutional neural network with VGG-19 model. In: 2021 IEEE World AI IoT Congress (AIIoT). IEEE; 2021.
Understanding and Coding a ResNet in Keras | by Priya Dwivedi | Towards Data Science. https://towardsdatascience.com/understanding-and-coding-a-resnet-in-keras-446d7ff84d33 (Accessed 28 Dec 2022).
Wen L, Li X, Gao L. A transfer convolutional neural network for fault diagnosis based on ResNet-50. Neural Comput Appl. 2020;32:6111–24.
Moses DA. Deep learning applied to automatic disease detection using chest X-rays. J Med Imaging Radiat Oncol. 2021;65(5):498–517. https://doi.org/10.1111/1754-9485.13273.
Kundu R, Das R, Geem ZW, Han GT, Sarkar R. Pneumonia detection in chest X-ray images using an ensemble of deep learning models. PLoS One. 2021;16(9): e0256630. https://doi.org/10.1371/JOURNAL.PONE.0256630.
Alharbi AH, Hosni Mahmoud HA. Pneumonia transfer learning deep learning model from segmented X-rays. Healthcare. 2022;10(6):987. https://doi.org/10.3390/HEALTHCARE10060987.
Niu S, et al. A decade survey of transfer learning (2010–2020). IEEE Trans Artif Intell. 2020;1(2):151–66.
Kumari S, Kumar D, Mittal M. An ensemble approach for classification and prediction of diabetes mellitus using soft voting classifier. Int J Cogn Comput Eng. 2021;2:40–6.
Sherazi SWA, Bae J-W, Lee JY. A soft voting ensemble classifier for early prediction and diagnosis of occurrences of major adverse cardiovascular events for STEMI and NSTEMI during 2-year follow-up in patients with acute coronary syndrome. PLoS One. 2021;16(6): e0249338.
1.11. Ensemble methods—scikit-learn 1.2.0 documentation. https://scikit-learn.org/stable/modules/ensemble.html (Accessed 28 Dec 2022).
How to develop voting ensembles with Python—MachineLearningMastery.com. https://machinelearningmastery.com/voting-ensembles-with-python/ (Accessed 28 Dec 2022).
Deep learning for image classification in Python with CNN. https://www.projectpro.io/article/deep-learning-for-image-classification-in-python-with-cnn/418.
Funding
No funding received.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
No conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Raju, N., Augustine, D.P. & Chandra, J. A Novel Artificial Intelligence System for the Prediction of Interstitial Lung Diseases. SN COMPUT. SCI. 5, 143 (2024). https://doi.org/10.1007/s42979-023-02524-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42979-023-02524-3