Abstract
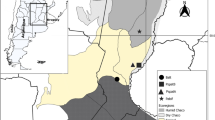
Bats can harbor a diversity of viruses, such as adenovirus. Ten different species of bat adenoviruses (BtAdV A to J) have been previous described worlwide. In Brazil, BtAdV was described in three species of phyllostomid species: Artibeus lituratus, Desmodus rotundus, and Sturnira lilium. There are around 180 bat species in Brazil, with 67% inhabiting the Atlantic Forest, with few information about the circulation of BtAdV in this biome. We aimed to describe the molecular detection and the phylogenetic characterization and suggest a classification of BtAdVs circulating in bats from the Brazilian Atlantic Forest. We collected 382 oral and rectal swabs from 208 bats between 2014–2015 and 2020–2021 from São Paulo, Pernambuco, and Santa Catarina Brazilian states. The adenovirus detection was done by a nested PCR targeting the DNA polymerase gene, and all positive samples were sequenced by the Sanger method. The phylogenetic analyses were based on the amino acid sequences using the MEGA 7 and BEAST software. We obtained 16 positive animals (detection rate 7.7%) belonging to seven bat species: Artibeus lituratus, Carollia perspicillata, Sturnira lilium, Molossus molossus, and the first record of Phyllostomus discolor, Eptesicus diminutus, and Myotis riparius. The phylogenetic analysis based on partial amino acid sequences showed that all obtained AdV sequences belong to the Mastadenovirus genus. We observed a high genetic diversity of BtAdV and identified eleven potential BtAdV species circulating in Brazil (BtAdV K to U). Our results contribute to the epidemiological surveillance of adenovirus, increasing the knowledge about the viral diversity and the distribution of AdV in bats from the Atlantic Forest.



Similar content being viewed by others
Data availability
The data supporting the findings of this study are available in the National Center for Biotechnology Information (NCBI) at https://www.ncbi.nlm.nih.gov. Additional data are available under reasonable request.
References
Davison AJ, Benkő M, Harrach B (2003) Genetic content and evolution of adenoviruses. J Gen Virol 84(Pt 11):2895–2908. https://doi.org/10.1099/vir.0.19497-0
Benkő M, Aoki K, Arnberg N, Davison AJ, Echavarría M, Hess M, Jones MS, Kaján GL, Kajon AE, Mittal SK, Podgorski II, San Martín C, Wadell G, Watanabe H, Harrach B, Ictv Report Consortium (2022) ICTV virus taxonomy profile: adenoviridae. J Gen Virol 103(3):001721. https://doi.org/10.1099/jgv.0.001721
Rex K, Kelm DH, Wiesner K, Kunz TH, Voigt CC (2008) Species richness and structure of three Neotropical bat assemblages. Biol J Linn Soc 94:12. https://doi.org/10.1111/j.1095-8312.2008.01014.x
Lau SK, Woo PC, Wong BH, Wong AY, Tsoi HW, Wang M, Lee P, Xu H, Poon RW, Guo R, Li KS, Chan KH, Zheng BJ, Yuen KY (2010) Identification and complete genome analysis of three novel paramyxoviruses, Tuhoko virus 1, 2 and 3, in fruit bats from China. Virology 404(1):106–116. https://doi.org/10.1016/j.virol.2010.03.049
Wang Y, Shen G, Hu D, Qian S, Zhu C, Tan W, Wang C (2017) Detection and identification of the bat circovirus BtCV-DS13. Bing Du Xue Bao 33(1):82–88
Leroy EM, Kumulungui B, Pourrut X, Rouquet P, Hassanin A, Yaba P, Délicat A, Paweska JT, Gonzalez JP, Swanepoel R (2005) Fruit bats as reservoirs of Ebola virus. Nature 438(7068):575–576. https://doi.org/10.1038/438575a
Tang XC, Zhang JX, Zhang SY, Wang P, Fan XH, Li LF, Li G, Dong BQ, Liu W, Cheung CL, Xu KM, Song WJ, Vijaykrishna D, Poon LL, Peiris JS, Smith GJ, Chen H, Guan Y (2006) Prevalence and genetic diversity of coronaviruses in bats from China. J Virol 80(15):7481–7490. https://doi.org/10.1128/JVI.00697-06
Calisher CH, Childs JE, Field HE, Holmes KV, Schountz T (2006) Bats: important reservoir hosts of emerging viruses. Clin Microbiol Rev 19(3):531–545. https://doi.org/10.1128/CMR.00017-06
Luis AD, Hayman DT, O'Shea TJ, Cryan PM, Gilbert AT, Pulliam JR, Mills JN, Timonin ME, Willis CK, Cunningham AA, Fooks AR, Rupprecht CE, Wood JL, Webb CT (2013) A comparison of bats and rodents as reservoirs of zoonotic viruses: are bats special? Proc Biol Sci 280(1756):20122753. https://doi.org/10.1098/rspb.2012.2753
Li Y, Ge X, Zhang H, Zhou P, Zhu Y, Zhang Y, Yuan J, Wang LF, Shi Z (2010) Host range, prevalence, and genetic diversity of adenoviruses in bats. J Virol 84(8):3889–3897. https://doi.org/10.1128/JVI.02497-09
Maeda K, Hondo E, Terakawa J, Kiso Y, Nakaichi N, Endoh D, Sakai K, Morikawa S, Mizutani T (2008) Isolation of novel adenovirus from fruit bat (Pteropus dasymallus yayeyamae). Emerg Infect Dis 14(2):347–349. https://doi.org/10.3201/eid1402.070932
Sonntag M, Mühldorfer K, Speck S, Wibbelt G, Kurth A (2009) New adenovirus in bats, Germany. Emerg Infect Dis 15(12):2052–2055. https://doi.org/10.3201/eid1512.090646
Han HJ, Wen HL, Zhao L, Liu JW, Luo LM, Zhou CM, Qin XR, Zhu YL, Liu MM, Qi R, Li WQ, Yu H, Yu XJ (2017) Novel coronaviruses, astroviruses, adenoviruses and circoviruses in insectivorous bats from northern China. Zoonoses Public Health 64(8):636–646. https://doi.org/10.1111/zph.12358
Ntumvi NF, Diffo JLD, Tamoufe U, Ndze VN, Takuo JM, Mouiche MMM, Nwobegahay J, LeBreton M, Gillis A, Rimoin AW, Schneider BS, Monagin C, McIver DJ, Joly DO, Wolfe ND, Rubin EM, Lange CE (2021) Evaluation of bat adenoviruses suggests co-evolution and host roosting behaviour as drivers for diversity. Microb Genom 7(4):000561. https://doi.org/10.1099/mgen.0.000561
Iglesias-Caballero M, Juste J, Vázquez-Morón S, Falcon A, Aznar-Lopez C, Ibáñez C, Pozo F, Ruiz G, Berciano JM, Garin I, Aihartza J, Echevarría JE, Casas I (2018) New adenovirus groups in Western Palaearctic bats. Viruses 10(8):443. https://doi.org/10.3390/v10080443
Vidovszky MZ, Kohl C, Boldogh S, Görföl T, Wibbelt G, Kurth A, Harrach B (2015) Random sampling of the Central European bat fauna reveals the existence of numerous hitherto unknown adenoviruses. Acta Vet Hung 63(4):508–525. https://doi.org/10.1556/004.2015.047
Hingst-Zaher E, Brandão MV (2021) Atlas Craniano: mamíferos da mata Atlântica e lista de espécies. Edições Tijd 10.32673/9786588932018
Abreu EF, Casali D, Costa-Araújo R, Garbino GST, Libardi GS, Loretto D, Loss AC, Marmontel M, Moras LM, Nascimento MC, Oliveira ML, Pavan SE, Tirelli FP (2022) Lista de Mamíferos do Brasil (2022-1). Zenodo. https://doi.org/10.5281/zenodo.7469767
Lima FE, Cibulski SP, Elesbao F, Carnieli Junior P, Batista HB, Roehe PM, Franco AC (2013) First detection of adenovirus in the vampire bat (Desmodus rotundus) in Brazil. Virus Genes 47(2):378–381. https://doi.org/10.1007/s11262-013-0947-6
Finoketti F, Santos RN, Campos AAS, Zani ALDS, Barboza CM, Fernandes MES, de Souza TCP, Santos DD, Bortolanza GW, Filho HO, Roehe PM, Franco AC, Batista HBCR (2019) Detection of adenovirus, papillomavirus and parvovirus in Brazilian bats of the species Artibeus lituratus and Sturnira lilium. Arch Virol 164(4):1015–1025. https://doi.org/10.1007/s00705-018-04129-1
PREDICT (2015) Adenoviridae: Adenovirus family project. https://www.ncbi.nlm.nih.gov/popset/1812582331?report=genbank. Accessed 19 Apr 2023
Wellehan JF, Johnson AJ, Harrach B, Benkö M, Pessier AP, Johnson CM, Garner MM, Childress A, Jacobson ER (2004) Detection and analysis of six lizard adenoviruses by consensus primer PCR provides further evidence of a reptilian origin for the atadenoviruses. J Virol 78(23):13366–13369. https://doi.org/10.1128/JVI.78.23.13366-13369.2004
Duvaud S, Gabella C, Lisacek F, Stockinger H, Ioannidis V, Durinx C (2021) Expasy, the Swiss Bioinformatics Resource Portal, as designed by its users. Nucleic Acids Res 49(W1):W216–W227. https://doi.org/10.1093/nar/gkab225
Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874. https://doi.org/10.1093/molbev/msw054
Suchard MA, Lemey P, Baele G, Ayres DL, Drummond AJ, Rambaut A (2018) Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol 4(1):vey016. https://doi.org/10.1093/ve/vey016
R Core Team (2021). R: a language and environment for statistical computing.
Jamovi (2021) The jamovi project. 2.2 edn.
IBGE (2022) Portal do IBGE - Instituto Brasileiro de Geografia e Estatística. https://www.ibge.gov.br/pt/inicio.html. Accessed 19 Apr 2023
Rossetto F, Iglesias-Caballero M, Liedtke HC, Gomez-Mestre I, Berciano JM, Pérez-Suárez G, de Paz O, Ibáñez C, Echevarría JE, Casas I, Juste J (2020) Mating strategy is determinant of adenovirus prevalence in European bats. PLoS One 15(1):e0226203. https://doi.org/10.1371/journal.pone.0226203
Zheng XY, Qiu M, Chen HF, Chen SW, Xiao JP, Jiang LN, Huo ST, Shi TL, Ma LZ, Liu S, Zhou JH, Zhang QH, Li X, Chen Z, Wu Y, Li JM, Guan WJ, Xiong YQ, Ma SJ et al (2016) Molecular detection and phylogenetic characterization of bat and human adenoviruses in Southern China. Vector Borne Zoonotic Dis 16(6):423–427. https://doi.org/10.1089/vbz.2015.1892
Jánoska M, Vidovszky M, Molnár V, Liptovszky M, Harrach B, Benko M (2011) Novel adenoviruses and herpesviruses detected in bats. Vet J 189(1):118–121. https://doi.org/10.1016/j.tvjl.2010.06.020
Hardmeier I, Aeberhard N, Qi W, Schoenbaechler K, Kraettli H, Hatt JM, Fraefel C, Kubacki J (2021) Metagenomic analysis of fecal and tissue samples from 18 endemic bat species in Switzerland revealed a diverse virus composition including potentially zoonotic viruses. PLoS One 16(6):e0252534. https://doi.org/10.1371/journal.pone.0252534
Waruhiu C, Ommeh S, Obanda V, Agwanda B, Gakuya F, Ge XY, Yang XL, Wu LJ, Zohaib A, Hu B, Shi ZL (2017) Molecular detection of viruses in Kenyan bats and discovery of novel astroviruses, caliciviruses and rotaviruses. Virol Sin 32(2):101–114. https://doi.org/10.1007/s12250-016-3930-2
Lee DN, Angiel M (2020) Two novel adenoviruses found in Cave Myotis bats (Myotis velifer) in Oklahoma. Virus Genes 56(1):99–103. https://doi.org/10.1007/s11262-019-01719-2
Anthony SJ, Epstein JH, Murray KA, Navarrete-Macias I, Zambrana-Torrelio CM, Solovyov A, Ojeda-Flores R, Arrigo NC, Islam A, Ali Khan S, Hosseini P, Bogich TL, Olival KJ, Sanchez-Leon MD, Karesh WB, Goldstein T, Luby SP, Morse SS, Mazet JA et al (2013) A strategy to estimate unknown viral diversity in mammals. mBio 4(5):e00598-00513. https://doi.org/10.1128/mBio.00598-13
Robinson CM, Singh G, Lee JY, Dehghan S, Rajaiya J, Liu EB, Yousuf MA, Betensky RA, Jones MS, Dyer DW, Seto D, Chodosh J (2013) Molecular evolution of human adenoviruses. Sci Rep 3:1812. https://doi.org/10.1038/srep01812
Risso-Ballester J, Cuevas JM, Sanjuán R (2016) Genome-wide estimation of the spontaneous mutation rate of human adenovirus 5 by high-fidelity deep sequencing. PLoS Pathog 12(11):e1006013. https://doi.org/10.1371/journal.ppat.1006013
Parker EJ, Botting CH, Webster A, Hay RT (1998) Adenovirus DNA polymerase: domain organisation and interaction with preterminal protein. Nucleic Acids Res 26(5):1240–1247. https://doi.org/10.1093/nar/26.5.1240
Uil TG, Vellinga J, de Vrij J, van den Hengel SK, Rabelink MJ, Cramer SJ, Eekels JJ, Ariyurek Y, van Galen M, Hoeben RC (2011) Directed adenovirus evolution using engineered mutator viral polymerases. Nucleic Acids Res 39(5):e30. https://doi.org/10.1093/nar/gkq1258
Tirera S, de Thoisy B, Donato D, Bouchier C, Lacoste V, Franc A, Lavergne A (2021) The influence of habitat on viral diversity in neotropical rodent hosts. Viruses 13(9):1690. https://doi.org/10.3390/v13091690
Muylaert RL, Matos DMS, Mello MAR (2014) Interindividual variations in fruit preferences of the yellow-shouldered bat Sturnira lilium (Chiroptera: Phyllostomidae) in a cafeteria experiment. Mammalia 78:9. https://doi.org/10.1515/mammalia-2012-0103
Acknowledgements
We thank the support of Brazilian Post and Telegraph Company (Correios) for transporting part of our field work supplies and samples. We thank Prof. Dr. Gislaine Fongaro from Department of Microbiology, Immunology and Parasitology, Federal University of Santa Catarina (UFSC) by human adenovirus 5 kindly provided. We thank the National Network for Virus Surveillance in Wild Animals (PREVIR-MCTI Network). We also thank Walm Engineering and Environmental Technology, which kindly authorized and enabled the collection of samples during fauna studies we performed in Joinville/SC.
Funding
This work is funded by the Ministry of Science, Technology, and Innovation (MCTI-Brazil) and by the National Council for Scientific and Technological Development (CNPq: 403761/2020-4 and 306595/2019-2). LMB is recipient of São Paulo Research Foundation (FAPESP) scholarship (2021/09144-1). LSR, LMNS, MVSM are recipient of Coordination for the Improvement of Higher Education Personnel (CAPES) scholarship, GPS, IBA, TO, INC, RDM, JCB, RCR, IMSP, LDNS, CWA, HLF are recipient of CNPq scholarship.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and performed the research. Data collection, analysis, and interpretation of data were performed by LSR, LMB, TCC, MVSM, AOV, LMNS, JCB, GPS, BLTL, IBA, TO, EGD, INC, RDM, GLL, RCR, IMSP, LDNS, EHZ, SMAJ, WRTJ, ELD, CWA, HLF. The first draft of the manuscript was written by LSR, LMB, TCC, HLF, and all authors commented on previous versions of the manuscript. The funding acquisition was done by ELD, CWA, and HLF. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
The authors confirm that the ethical policies of the journal, as noted in the journal’s author guidelines page, have been adhered to, and the appropriate ethical review committee approval has been received. All applicable international, national, and institutional guidelines for the care and use of animals were followed. All the sample collections were authorized by the Brazilian Institute of Environment and Natural Renewable Resources (SISBIO: 25895, 76621 and 45807-2) and by the Ethics Committee on the Use of Animals of the University of São Paulo (CEUA 1128141120 and CEUAx 7141081019).
Competing interests
The authors declare no competing interests.
Additional information
Responsible Editor: Fernando R. Spilki
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary file 1
Table S1 Summary of obtained samples in this study: Genbank number, ID number, bat species, bat family, age, Brazilian location, collection date, and % amino acid identity (DOCX 16 kb)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Rizotto, L.S., Bueno, L.M., Corrêa, T.C. et al. Genetic diversity of adenovirus in neotropical bats from Brazil. Braz J Microbiol 54, 3221–3230 (2023). https://doi.org/10.1007/s42770-023-01109-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42770-023-01109-9