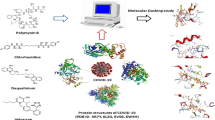
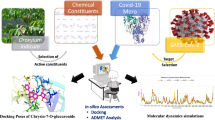
Abstract
The 2019-nCoV virus is a human-infectious coronavirus (CoV). The healthcare workers fighting this outbreak on the front lines have few treatment options available. The main warning symptoms of COVID-19, the disease caused by the new coronavirus, are fever, fatigue, and a dry cough, and it also causes cold-like symptoms like a runny nose which are sometimes similar to symptoms of allergies and sometimes difficult to differentiate between COVID-19 and allergies. The anti-allergic drug molecules can behave as good inhibitors against COVID-19. Molecular docking studies have been performed to examine the inhibitor properties of anti-allergic molecules against COVID-19. The search for better inhibitors has been examined interns of various non-covalent interactions like hydrogen bonds, halogen bonds, van der Waals interactions, and alkyl-n and n–n interactions between small molecules (anti-allergic medicines) with the main protease of COVID-19 using molecular docking and molecular dynamics simulation which reveals that astemizole is the best inhibitor among ten anti-allergic drug molecules.
Graphical abstract
















Similar content being viewed by others
References
Allergy Symptoms vs. Coronavirus (COVID-19), WebMD, https://www.webmd.com/lung/covid-allergies
Al-Osai AM, Al-Wazzah MJ (2017) The history and epidemiology of Middle East respiratory syndrome corona virus. Multidiscip Respir Med 12:20. https://doi.org/10.1186/s40248-017-0101-8
Baildya N, Ghosh NN, Chattopadhyay AP (2020) Inhibitory activity of hydroxychloroquine on COVID-19 main protease: an insight from MD-simulation studies. J Mol Struct 5(1219):128595
Baranov P, Henderson VCM, Anderson CB, Gesteland RF, Atkins JF, Howard MT (2005) Programmed ribosomal frameshifting in decoding the SARS-CoV genome. Virology 332:498–510
Berendsen HJC, Postma JPM, Van Gunsteren WF, DiNola A, Haak JR (1984) Molecular dynamics with coupling to an external bath. J Chem Phys 81:3684–3690
Berman H, Battistuz MT, Bhat TN, Bluhm WF, Bourne PE, Burkhardt K, Feng Z, Gilliland GL, Iype L, Jain S et al (2002) The protein data bank. Acta Crystallogr Sect d: Biol Crystallogr 58:899–907
Bussi G, Donadio D, Parrinello M (2007) Canonical sampling through velocity rescaling. J Chem Phys 126:014101
Dallakyan S, Olson AJ (2016) Small molecule library screening by docking with PyRx. Methods Mol Biol 1263:243–250
Dassault Systemes, BIOVIA (2016) Discovery studio modeling environment, Release 2017, San Diego: Dassault Systemes
Dong N, Yang X, Ye L, Chen K, Chan E, Yang M, Chen S (2020) Genomic and protein structure modeling analysis depicts the origin and infectivity of 2019-nCoV, a new coronavirus which caused a pneumonia outbreak in Wuhan, China. bioRxiv. https://doi.org/10.1101/2020.01.20.913368v2
Fantini J, Di Scala C, Chahinian H, Yahi N (2020) Structural and molecular modelling studies reveal a new mechanism of action of chloroquine and hydroxychloroquine against SARS-CoV-2 infection. Int J Antimicrob Agents 55:105960
Ferron F, Subissi L, Silveira De Morais AT, Le NTT, Sevajol M, Gluais L et al (2018) Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc Natl Acad Sci 115:E162–E171
Furuta Y, Takahashi K, Shiraki K, Sakamoto K, Smee D, Barnard DL et al (2009) T-705 (favipiravir) and related compounds: Novel broad-spectrum inhibitors of RNA viral infections. Antivir Res 82:95–102
Gralinski L, Menachery V (2020) Return of the Coronavirus: 2019-nCoV. Viruses 24:135
Hall DC Jr, Ji HF (2020) A search for medications to treat COVID-19 via in silico molecular docking models of the SARS-CoV-2 spike glycoprotein and 3CL protease. Travel Med Infect Dis 35:101646
Krajczyk A, Kulinska K, Kulinski T, Hurst BL, Day CW, Smee DF et al (2014) Antivirally active ribavirin analogues—4,5-disubstituted 1,2,3-triazole nucleosides: biological evaluation against certain respiratory viruses and computational modelling. Antivir Chem Chemother 23:161–171
Li F, Li W, Farzan M, Harrison SC (2005) Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science 309:1864–1868
Li CC, Wang XJ, Wang HCR (2019) Repurposing host-based therapeutics to control coronavirus and influenza virus. Drug Discov Today 24:726–736
Liang C, Tian L, Liu Y, Hui N, Qiao G, Li H (2020) A promising antiviral candidate drug for the COVID-19 pandemic: a mini-review of remdesivir. Eur J Med Chem 201:112527
Liu X, Zhang B, Jin Z, Yang H, Rao Z (2020) The crystal structure of COVID-19 main protease in complex with an inhibitor N3. Protein DataBank. https://doi.org/10.2210/pdb6LU7/pdb
Lu H (2020) Drug treatment options for the 2019-new coronavirus (2019-nCoV). Biosci Trends 14:69–71
Morse JS, Lalonde T, Xu S, Liu WR (2020) Learning from the past: possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019-nCoV. Chembiochem 21(5):730–738. https://doi.org/10.1002/cbic.202000047
Pepperrell T, Pilkington V, Owen A, Wang J, Hill AM (2020) Review of safety and minimum pricing of nitazoxanide for potential treatment of COVID-19. J Virus Erad 6:52–60
Prajapat M, Sarma P, Shekhar N, Avti P, Sinha S, Kaur H, Kumar S, Bhattacharyya A, Kumar H, Bansal S, Medhi B (2020) Drug targets for corona virus: a systematic review. Indian J Pharmacol 52:56–65
Roosa S, Tikkanen MPH, Res M, Eric C, Schneider MD (2021) Social spending to improve population health — does the United States spend as wisely as other countries? N Engl J Med 382:885–887
Shi D, Wheelhouse DS, Hurley LH (2001) Quadruplex-interactive agents as telomerase inhibitors: synthesis of porphyrins and structure-activity relationship for the inhibition of telomerase. J Med Chem 44:4509–4523
Shoemaker QB, Thiessen A, Yu PA (2019) PubChem 2019 update: improved access to chemical data. Nucleic Acids Res 47:D1102–D1109
Si MK, Sen A, Ganguly B (2017) Exploiting hydrogen bonding interactions to probe smaller linear and cyclic diamines binding to G-quadruplexes: a DFT and molecular dynamics study. J Phys Chem Chem Phys 19:11474
Spezzani V, Piunno A, Iselin H (2020) Benign COVID-19 in an immunocompromised cancer patient—the case of a married couple. Swiss Med Wkly 150:w20246
Tilmanis DC, Van Baalen OhDY, Rossignol JF, Hurt AC (2017) The susceptibility of circulating human influenza viruses to tizoxanide, the active metabolite of nitazoxanide. Antivir Res 147:142–148
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31:455–461
Wakchaure PD, Velayutham R, Roy KK (2019) Structure investigation, enrichment analysis and structure-based repurposing of FDA-approved drugs as inhibitors of BET-BRD4. J Biomol Struct Dyn 37:3048–3057
Wakchaure PD, Ghosh S, Ganguly B (2020) Revealing the inhibition mechanism of RNA- dependent RNA polymerase (RdRp) of SARS-CoV-2 by remdesivir and nucleotide analogues: a molecular dynamics simulation study. J Phys Chem B 124:10641–10652
Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FTT, De Beer AP, Rempfer C, Bordoli L et al (2018) SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46:W296–W303
Xu X, Liu Y, Weiss S, Arnold E, Sarafianos SG, Ding J (2003) Molecular model of SARS coronavirus polymerase: implications for biochemical functions and drug design. Nucleic Acids Res 31:7117–7130
Zhang L, Zhou R (2020) Structural basis of the potential binding mechanism of remdesivir to SARS-CoV-2 RNA-dependent RNA polymerase. J Phys Chem B 124:6955–6962
Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen JY, Luo GH, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao G, Shi ZA (2020) Pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273
Zhu Z, Lu Z, Xu T, Chen C, Yang G, Zha T et al (2020) Arbidol monotherapy is superior to lopinavir/ritonavir in treating COVID-19. J Infect 81:e21–e23
Ziebuhr J, Snijder EJ, Gorbalenya AE (2000) Virus-encoded proteinases and proteolytic processing in the Nidovirales. J Gen Virol 81:853–879
Acknowledgements
The authors are thankful to Sardar Patel University, Balaghat, M.P, India & D. B. Science College, Gondia, India for facilitating our research work. The authors are also thankful to Dr. Kalyanashis Jana CSIR-CSMCRI for their support.
Author information
Authors and Affiliations
Contributions
MKS and MRP wrote the main manuscript text and SP prepared the figures. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Si, M.K., Patle, M.R. & Pandey, S. Molecular docking and molecular dynamics studies of anti-allergic medicines as inhibitors against COVID-19. J Proteins Proteom 14, 263–276 (2023). https://doi.org/10.1007/s42485-023-00122-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42485-023-00122-8