Abstract
We consider verifying the effect of the treatment under the situation in which a response is given when a treatment is applied to units with features. In estimating the effect, there are problems such as the treatment can be given only once to the unit and the features of the unit cannot be controlled. For such problems, conventional studies have some mathematical models. However, in this paper, we propose the different data generative model in which there are latent classes of units with the same response, each latent class contains units with similar features. We call this model the identical response structure latent class model (IRSLC model). Under the proposed model, we calculate the Bayes optimal decision and its approximation algorithm for the difference with and without the treatment for the entire population. We conducted experiments using the synthetic data of the model assumed by the proposed method or the conventional method. Then, we compared our method with previous studies to confirm the characteristics of the proposed model.







Similar content being viewed by others
References
Berger, J.O.: Statistical decision theory and Bayesian analysis. Springer, UK (1985)
Bishop, C.M.: Pattern recognition and machine learning. Springer, UK (2006)
Chen, H., Harinen, T., Lee, J.-Y., Yung, M., Z, Zhao.: Causalml: Python package for causal machine learning, (2020)
Imbens, G.W., Rubin, D.B.: Causal Inference for Statistics, Social, and Biomedical Sciences: An Introduction. Cambridge University Press, UK (2015)
Ishiwatari, T., Saito, S., Nakahara, Y., Iikubo, Y., Matsushima, T.: Bayes optimal estimation and its approximation algorithm for difference with and without treatment under URLC model, International Symposium on Information Theory and Its Applications, (2022)
Rudas, S.J.K.: Linear regression for uplift modeling. Data Min. Knowl. Discovery 32(5), 1275–1305 (2018)
Murayama, H., Saito, S., Iikubo, Y., Nakahara, Y., Matsushima, T.: Cluster’s number free bayes prediction of general framework on mixture of regression models. J. Stat. Theory Appl. 20, 425–449 (2021)
Pearl, J.: Causal diagrams for empirical research. Biometrika 82(4), 669–710 (1995)
Rosenbaum, P.R., Rubin, D.B.: The central role of the propensity score in observational studies for causal effects. Biometrika 70(1), 41–55 (1983)
Sharma, A., Kiciman E.: et al. DoWhy: A Python package for causal inference. https://github.com/microsoft/dowhy, (2019)
Acknowledgements
This work was supported in part by JSPS KAKENHI Grant Numbers JP17K06446, JP19K04914, JP19K14989, JP22K02811, and JP22K14254.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix A: Proof of proposition 2
Appendix A: Proof of proposition 2
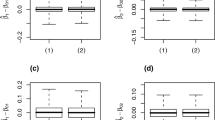
By substituting the approximate posterior distribution \(q^{(T)}\)\(({\varvec{z}}_k^N, {\varvec{\theta }}_k, k) =q^{(T)}(k) q^{(T)}({\varvec{z}}^N_k,{\varvec{\theta }}_k\mid k)\) in the posterior distribution \(p({\varvec{z}}_k^N,{\varvec{\theta }}_k,k\mid {\varvec{D}})\), we have
To calculate (A5), we define
Then, we can rewrite (A5) as
where \((\check{\beta }_{w{l_k,1}}^{(T)},\dots , \check{\beta }_{w{l_k,p+1}}^{(T)})\) are elements of \(\check{{\varvec{\beta }}}_{w{l_k}}^{(T)}\) and
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ishiwatari, T., Saito, S., Nakahara, Y. et al. Bayes optimal estimation and its approximation algorithm for difference with and without treatment under IRSLC model. Int J Data Sci Anal (2023). https://doi.org/10.1007/s41060-023-00468-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s41060-023-00468-8