Abstract
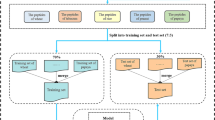
Protein 2-hydroxyisobutyrylation (Khib), a newly identified post-translational modification, plays a role in various cellular processes. To gain a comprehensive understanding of its regulatory mechanisms, it is crucial to identify the sites of 2-hydroxyisobutyrylation. Therefore, we developed a novel ensemble method, DeepKPred, for predicting species-specific 2-hydroxyisobutyrylation sites. We employed one-hot and AAindex encoding schemes to construct features from protein sequences and integrated two densely convolutional neural networks and two long short-term memory networks to build the model. In the 5-fold cross-validation dataset, DeepKPred achieved AUC values of 0.859, 0.804, 0.821, and 0.819 for Human, Candida albicans, Rice, Wheat, and Physcomitrella patens. Additionally, function analysis further indicated that different organisms tend to engage in distinct biological processes and pathways. Detailed analysis can help us learn more about the mechanism of 2-hydroxyisobutyrylation and provide insights for associated experimental verification.






Similar content being viewed by others
References
Shi Y (2022) Advances in big data analytics: theory, algorithms and practices. Springer, Singapore
Olson DL (2005) Introduction to business data mining. McGraw-Hill/Irwin, New York
Shi Y, Tian YJ, Kou G, Peng Y, Li J (2011) Optimization based data mining: theory and applications. Springer, Berlin
Tien JM (2017) Internet of things, real-time decision making, and Artificial Intelligence. Ann Data Sci 4:149–178
Walsh CT, Garneau-Tsodikova S, Gatto GJ Jr (2005) Protein posttranslational modifications: the chemistry of proteome diversifications. Angew Chem Int Ed Engl 44(45):7342–7372
Filtz TM, Vogel WK, Leid M (2014) Regulation of transcription factor activity by interconnected post-translational modifications. Trends Pharmacol Sci 35(2):76–85
Consortium U (2019) UniProt: a worldwide hub of protein knowledge. Nucl Acids Res 47(D1):506–515
Zhang W, Tan X, Lin S, Gou Y, Han C, Zhang C, Ning W, Wang C, Xue Y (2022) CPLM 4.0: an updated database with rich annotations for protein lysine modifications. Nucl Acids Res 50(D1):451–459
Dai L, Peng C, Montellier E, Lu Z, Chen Y, Ishii H, Debernardi A, Buchou T, Rousseaux S, Jin F, Sabari BR, Deng Z, Allis CD, Ren B, Khochbin S, Zhao Y (2014) Lysine 2-hydroxyisobutyrylation is a widely distributed active histone mark. Nat Chem Biol 10(5):365–370
Huang H, Tang S, Ji M, Tang Z, Shimada M, Liu X, Qi S, Locasale JW, Roeder RG, Zhao Y, Li X (2018) p300-Mediated lysine 2-Hydroxyisobutyrylation regulates glycolysis. Mol Cell 70(4):663–678e666
Huang J, Luo Z, Ying W, Cao Q, Huang H, Dong J, Wu Q, Zhao Y, Qian X, Dai J (2017) 2-Hydroxyisobutyrylation on histone H4K8 is regulated by glucose homeostasis in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 114(33):8782–8787
Huang S, Tang D, Dai Y (2020) Metabolic functions of lysine 2-Hydroxyisobutyrylation. Cureus 12(8):e9651
Qi T, Li J, Wang H, Han X, Li J, Du J (2021) Global analysis of protein lysine 2-hydroxyisobutyrylation (Khib) profiles in Chinese herb rhubarb (Dahuang). BMC Genomics 22(1):542
Umlauf D, Goto Y, Feil R (2004) Site-specific analysis of histone methylation and acetylation. Methods Mol Biol 287:99–120
Agarwal KL, Kenner GW, Sheppard RC (1969) Feline gastrin. An example of peptide sequence analysis by mass spectrometry. J Am Chem Soc 91(11):3096–3097
Medzihradszky KF (2005) Peptide sequence analysis. Methods Enzymol 402:209–244
Tian Y, Fu S (2020) A descriptive framework for the field of deep learning applications in medical images. Knowl Based Syst 210:106445
Ju Z, Wang SY (2019) iLys-Khib: identify lysine 2-hydroxyisobutyrylation sites using mRMR feature selection and fuzzy SVM algorithm. Chemometr Intell Lab Syst 191:96–102
Wang YG, Huang SY, Wang LN, Zhou ZY, Qiu JD (2020) Accurate prediction of species-specific 2-hydroxyisobutyrylation sites based on machine learning frameworks. Anal Biochem 602:113793
Zhang L, Zou Y, He N, Chen Y, Chen Z, Li L (2020) DeepKhib: a deep-learning framework for lysine 2-hydroxyisobutyrylation sites prediction. Front Cell Dev Biol 8:580217
Jia X, Zhao P, Li F, Qin Z, Ren H, Li J, Miao C, Zhao Q, Akutsu T, Dou G, Chen Z, Song J (2023) ResNetKhib: a novel cell type-specific tool for predicting lysine 2-hydroxyisobutylation sites via transfer learning. Brief Bioinform 24(2). https://doi.org/10.1093/bib/bbad063
Wu Q, Ke L, Wang C, Fan P, Wu Z, Xu X (2018) Global analysis of lysine 2-hydroxyisobutyrylome upon SAHA treatment and its relationship with acetylation and crotonylation. J Proteome Res 17(9):3176–3183
Zheng H, Song N, Zhou X, Mei H, Li D, Li X, Liu W (2021) Proteome-wide analysis of lysine 2-Hydroxyisobutyrylation in Candida albicans. mSystems 6(1):10–1128. https://doi.org/10.1128/mSystems.01129-20
Meng X, Xing S, Perez LM, Peng X, Zhao Q, Redoña ED, Wang C, Peng Z (2017) Proteome-wide analysis of lysine 2-hydroxyisobutyrylation in developing Rice (Oryza sativa) seeds. Sci Rep 7(1):17486
Yu Z, Ni J, Sheng W, Wang Z, Wu Y (2017) Proteome-wide identification of lysine 2-hydroxyisobutyrylation reveals conserved and novel histone modifications in Physcomitrella patens. Sci Rep 7(1):15553
Bo F, Shengdong L, Zongshuai W, Fang C, Zheng W, Chunhua G, Geng L, Ling’an K (2021) Global analysis of lysine 2-hydroxyisobutyrylation in wheat root. Sci Rep 11(1):6327
Kawashima S, Pokarowski P, Pokarowska M, Kolinski A, Katayama T, Kanehisa M (2008) AAindex: amino acid index database, progress report 2008. Nucl Acids Res 36:202–205. https://doi.org/10.1093/nar/gkm998
Lin CT, Lin KL, Yang CH, Chung IF, Huang CD, Yang YS (2005) Protein metal binding residue prediction based on neural networks. Int J Neural Syst 15(1–2):71–84
Hochreiter S, Schmidhuber J (1997) Long short-term memory. Neural Comput 9(8):1735–1780
Fukushima K (1980) Neocognitron: a self organizing neural network model for a mechanism of pattern recognition unaffected by shift in position. Biol Cybern 36(4):193–202
Zhang Z, Xie H, Zuo W, Tang J, Zeng Z, Cai W, Lai L, Lu Y, Shen L, Dong X, Yin L, Tang D, Dai Y (2021) Lysine 2-hydroxyisobutyrylation proteomics reveals protein modification alteration in the actin cytoskeleton pathway of oral squamous cell carcinoma. J Proteom 249:104371
Vacic V, Iakoucheva LM, Radivojac P (2006) Two sample logo: a graphical representation of the differences between two sets of sequence alignments. Bioinformatics 22(12):1536–1537
Funding
This work is supported by grants from the Natural Science Foundation of China (12071024) and the statistics and materials interdisciplinary Project No.00003957.
Author information
Authors and Affiliations
Contributions
YX conceived and designed the experiments. SF performed the experiments and wrote the paper. YX. revised the manuscript. All the authors read and agreed on the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing financial interests.
Ethical statement
I hereby declare that this manuscript is the result of our (Shiqi Fan and Yan Xu) independent creation under the reviewers’ comments. Except for the quoted contents, this manuscript does not contain any research achievements that have been published or written by other individuals or groups. I am the corresponding author of this manuscript. The legal responsibility of the statement shall be borne by me.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Fan, S., Xu, Y. DeepKPred: Prediction and Functional Analysis of Lysine 2-Hydroxyisobutyrylation Sites Based on Deep Learning. Ann. Data. Sci. 11, 693–707 (2024). https://doi.org/10.1007/s40745-023-00504-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40745-023-00504-1