Abstract
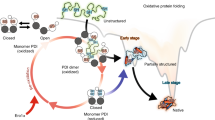
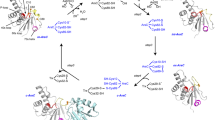
The dynamic behavior of Protein Disulfide Isomerase (PDI) in an aqueous solution environment under physiologically active pH has been experimentally verified in this study using Small Angle X-ray Scattering (SAXS) technique. The structural mechanism of dimerization for full-length PDI molecules and co-complex with two renowned substrates has been comprehensively discussed. The structure models obtained from the SAXS data of PDI purified from bovine liver display behavior duality between unaccompanied-enzyme and after engaged with substrates. The analysis of SAXS data revealed that PDI exists as a homo-dimer in the solution environment, and substrate induction provoked its segregation into monomer to enable the enzyme to interact systematically with incoming clients.










Similar content being viewed by others
Data avilability
The Small Angle Scattering Biological Data Bank (SASBDB). Editors can access the depositions at the following URLs: https://www.sasbdb.org/data/SASDGZ9/j3szubgaha/https://www.sasbdb.org/data/SASDH22/ngwcjmyyj2/https://www.sasbdb.org/data/SASDH32/za9s4e66vu/https://www.sasbdb.org/data/SASDH42/en5k9t9532/https://www.sasbdb.org/data/SASDH52/gu0rrywqqf/. A full project summary can be located here: https://www.sasbdb.org/project/908/9er70jdgos/.
Abbreviations
- PDI:
-
Protein Disulfide Isomerase
- yPDI:
-
Yeast PDI
- hPDI:
-
Human PDI
- SDS:
-
Sodium dodecyl sulfate
- PAGE:
-
Polyacrylamide gel electrophoresis
- SAXS:
-
Small-angle X-ray scattering
- PDB:
-
Protein data bank
- BPTI:
-
Bovine pancreatic trypsin inhibitor
- RNase:
-
Ribonuclease
- BS-RNase:
-
Bull seminal ribonuclease
- TCEP:
-
Tris(2-carboxyethyl)phosphine
- 1 M:
-
One molemM: millimolekDa: kilo Dalton
- EDTA:
-
Ethylene diamine tetraacetic acid;
- NaCl:
-
Sodium chlorideDTT: Dithiothreitol
- α1:
-
Alpha helix 1
- α3:
-
Alpha helix 3
- α4:
-
Alpha helix 4
- β4:
-
Beta-strand 4
References
Bastos-Aristizabal S, Kozlov G, Gehring K (2014) Structural insight into the dimerization of human protein disulfide isomerase. Protein Sci 23:618–626. https://doi.org/10.1002/pro.2444
Benham AM, van Lith M, Sitia R, Braakman I (2013) Ero1-PDI interactions, the response to redox flux and the implications for disulfide bond formation in the mammalian endoplasmic reticulum. Philos Trans R Soc Lond B Biol Sci 368:20110403. https://doi.org/10.1098/rstb.2011.0403
Bottomley MJ, Batten MR, Lumb RA, Bulleid NJ (2001) Quality control in the endoplasmic reticulum: PDI mediates the ER retention of unassembled procollagen C- propeptides. Curr Biol 11:1114–1118. https://doi.org/10.1016/S0960-9822(01)00317-7
Byrne LJ, Sidhu A, Wallis AK, Ruddock LW, Freedman RB, Howard MJ, Williamson RA (2009) Mapping of the ligand-binding site on the b’ domain of human PDI: interaction with peptide ligands and the x-linker region. Biochem J 423:209–217. https://doi.org/10.1042/BJ20090565
Carmichael DF, Morin JE, Dixon JE (1977) Purification and characterization of a thiol:protein disulfide oxidoreductase from bovine liver. J Biol Chem 252:7163–7167
Chatani E, Hayashi R, Moriyama H, Ueki T (2002) Conformational strictness required for maximum activity and stability of bovine pancreatic ribonuclease A as revealed by crystallographic study of three Phe120 mutants at 1.4 A resolution. Protein Sci 11:72–81. https://doi.org/10.1110/ps.31102
Cherubin P, Guyette J, Taylor M, O’Donnell M, Herndon L, Burress H, Riad A, Tatulian SA, Teter K (2018) Protein disulfide isomerase does not act as an unfoldase in the disassembly of cholera toxin. Biosci Rep. https://doi.org/10.1042/BSR20181320
Dunne M, Leicht S, Krichel B, Mertens HDT, Thompson A, Krijgsveld J, Svergun DI, Gómez-Torres N, Garde S, Uetrecht C, Narbad A, Mayer MJ, Meijers R (2016) Crystal structure of the CTP1L endolysin reveals how its activity is regulated by a secondary translation product. J Biol Chem 291:4882–4893. https://doi.org/10.1074/jbc.M115.671172
Franke D, Svergun DI (2009) DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J Appl Crystallogr 42:342–346. https://doi.org/10.1107/S0021889809000338
Franke D, Kikhney AG, Svergun DI (2012) Automated acquisition and analysis of small angle X-ray scattering data. Nucl Instrum Methods Phys Res Sect A Accel Spectrometers Detect Assoc Equip 689:52–59. https://doi.org/10.1016/j.nima.2012.06.008
Franke D, Petoukhov MV, Konarev PV, Panjkovich A, Tuukkanen A, Mertens HDT, Kikhney AG, Hajizadeh NR, Franklin JM, Jeffries CM, Svergun DI (2017) ATSAS 2.8: a comprehensive data analysis suite for small-angle scattering from macromolecular solutions. J Appl Crystallogr 50:1212–1225. https://doi.org/10.1107/S1600576717007786
Goldberger RF, Epstein CJ, Anfinsen CB (1963) Acceleration of reactivation of reduced bovine pancreatic ribonuclease by a microsomal system from rat liver. J Biol Chem 238:628–635
Gonzalez V, Pal R, Narayan M (2010) The oxidoreductase behavior of protein disulfide isomerase impedes fold maturation of endoplasmic reticulum-processed proteins in the pivotal structure-coupled step of oxidative folding: implications for subcellular protein trafficking. Biochemistry 49:6282–6289. https://doi.org/10.1021/bi100753s
Gruber CW, Cemazar M, Heras B, Martin JL, Craik DJ (2006) Protein disulfide isomerase: the structure of oxidative folding. Trends Biochem Sci 31:455–464. https://doi.org/10.1016/j.tibs.2006.06.001
Hatahet F, Ruddock LW (2009) Protein disulfide isomerase: a critical evaluation of its function in disulfide bond formation. Antioxid Redox Signal 11:2807–2850. https://doi.org/10.1089/ars.2009.2466
Heidorn DB, Trewhella J (1988) Comparison of the crystal and solution structures of calmodulin and troponin C. Biochemistry 27:909–915. https://doi.org/10.1021/bi00403a011
Hu CH, Tsou CL (1992) C-terminal truncation of bovine protein disulfide isomerase increases its activity. Biochem Biophys Res Commun 183:714–718. https://doi.org/10.1016/0006-291X(92)90541-R
Koivunen P, Salo KEH, Myllyharju J, Ruddock LW (2005) Three binding sites in protein- disulfide isomerase cooperate in collagen prolyl 4-hydroxylase tetramer assembly. J Biol Chem 280:5227–5235. https://doi.org/10.1074/jbc.M412480200
Kozin MB, Svergun DI (2001) Automated matching of high- and low-resolution structural models. J Appl Crystallogr 34:33–41. https://doi.org/10.1107/S0021889800014126
Kulp MS, Frickel E-M, Ellgaard L, Weissman JS (2006) Domain architecture of protein- disulfide isomerase facilitates its dual role as an oxidase and an isomerase in Ero1p- mediated disulfide formation. J Biol Chem 281:876–884. https://doi.org/10.1074/jbc.M511764200
Lambert N, Freedman RB (1983) Structural properties of homogeneous protein disulphide- isomerase from bovine liver purified by a rapid high-yielding procedure. Biochem J 213:225–234. https://doi.org/10.1042/bj2130225
Li S, Hong X, Shi Y, Li H, Wang C (2006) Annular arrangement and collaborative actions of four domains of protein-disulfide isomerase: a small angle X-ray scattering study in solution. J Biol Chem 281:6581–6588. https://doi.org/10.1074/jbc.M508422200
Li H, Yang K, Wang W, Niu Y, Li J, Dong Y, Liu Y, Wang C-C, Wang L, Liang H (2018) Crystal and solution structures of human protein-disulfide isomerase-like protein of the testis (PDILT) provide insight into its chaperone activity. J Biol Chem 293:1192–1202. https://doi.org/10.1074/jbc.M117.797290
Liu Y, Hart PJ, Schlunegger MP, Eisenberg D (1998) The crystal structure of a 3D domain- swapped dimer of RNase A at a 21-A resolution. Proc Natl Acad Sci U S a 95:3437–3442. https://doi.org/10.1073/pnas.95.7.3437
Lyles MM, Gilbert HF (1991) Catalysis of the oxidative folding of ribonuclease A by protein disulfide isomerase: dependence of the rate on the composition of the redox buffer. Biochemistry 30:613–619. https://doi.org/10.1021/bi00217a004
Mazzarella L, Capasso S, Demasi D, Di Lorenzo G, Mattia CA, Zagari A (1998) Bovine seminal ribonuclease: structure at 1.9 A resolution. Acta Crystallogr D Biol Crystallogr 49(1993):389–402. https://doi.org/10.1107/S0907444993003403
Morjana NA, McKeone BJ, Gilbert HF (1993) Guanidine hydrochloride stabilization of a partially unfolded intermediate during the reversible denaturation of protein disulfide isomerase. Proc Natl Acad Sci U S a 90:2107–2111
Nakasako M, Maeno A, Kurimoto E, Harada T, Yamaguchi Y, Oka T, Takayama Y, Iwata A, Kato K (2010) Redox-dependent domain rearrangement of protein disulfide isomerase from a thermophilic fungus. Biochemistry 49:6953–6962. https://doi.org/10.1021/bi1006089
Okumura M, Noi K, Kanemura S, Kinoshita M, Saio T, Inoue Y, Hikima T, Akiyama S, Ogura T, Inaba K (2019) Dynamic assembly of protein disulfide isomerase in catalysis of oxidative folding. Nat Chem Biol 15:499–509. https://doi.org/10.1038/s41589-019-0268-8
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF chimera–a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. https://doi.org/10.1002/jcc.20084
Pihlajaniemi T, Helaakoski T, Tasanen K, Myllylä R, Huhtala ML, Koivu J, Kivirikko KI (1987) Molecular cloning of the beta-subunit of human prolyl 4-hydroxylase. This subunit and protein disulphide isomerase are products of the same gene. EMBO J 6:643–649
Pirneskoski A, Klappa P, Lobell M, Williamson RA, Byrne L, Alanen HI, Salo KEH, Kivirikko KI, Freedman RB, Ruddock LW (2004) Molecular characterization of the principal substrate binding site of the ubiquitous folding catalyst protein disulfide isomerase. J Biol Chem 279:10374–10381. https://doi.org/10.1074/jbc.M312193200
Schwaller M, Wilkinson B, Gilbert HF (2003) Reduction-reoxidation cycles contribute to catalysis of disulfide isomerization by protein-disulfide isomerase. J Biol Chem 278:7154–7159. https://doi.org/10.1074/jbc.M211036200
Seaton BA, Head JF, Engelman DM, Richards FM (1985) Calcium-induced increase in the radius of gyration and maximum dimension of calmodulin measured by small-angle X-ray scattering. Biochemistry 24:6740–6743. https://doi.org/10.1021/bi00345a002
Severino A, Campioni M, Straino S, Salloum FN, Schmidt N, Herbrand U, Frede S, Toietta G, Di Rocco G et al (2007) Identification of protein disulfide isomerase as a cardiomyocyte survival factor in ischemic cardiomyopathy. J Am Coll Cardiol 50:1029–1037. https://doi.org/10.1016/j.jacc.2007.06.006
Smith AM, Chan J, Oksenberg D, Urfer R, Wexler DS, Ow A, Gao L, McAlorum A, Huang S-G (2004) A high-throughput turbidometric assay for screening inhibitors of protein disulfide isomerase activity. J Biomol Screen 9:614–620. https://doi.org/10.1177/1087057104265292
Solovyov A, Gilbert HF (2004) Zinc-dependent dimerization of the folding catalyst, proteindisulfideisomerase. ProteinSci 3:1902–1907. https://doi.org/10.1110/ps.04716104
Svergun DI (1999) Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys J 76:2879–2886. https://doi.org/10.1016/S0006-3495(99)77443-6
Svergun D, Barberato C, Koch MHJ (1995) CRYSOL ? a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Crystallogr 28:768–773. https://doi.org/10.1107/S0021889895007047
Taylor M, Burress H, Banerjee T, Ray S, Curtis D, Tatulian SA, Teter K (2014) Substrate- induced unfolding of protein disulfide isomerase displaces the cholera toxin A1 subunit from its holotoxin. PLoS Pathog 10:e1003925. https://doi.org/10.1371/journal.ppat.1003925
Tian G, Xiang S, Noiva R, Lennarz WJ, Schindelin H (2006) The crystal structure of yeast protein disulfide isomerase suggests cooperativity between its active sites. Cell 124:61–73. https://doi.org/10.1016/j.cell.2005.10.044
Tian G, Kober F-X, Lewandrowski U, Sickmann A, Lennarz WJ, Schindelin H (2008) The catalytic activity of protein-disulfide isomerase requires a conformationally flexible molecule. J Biol Chem 283:33630–33640. https://doi.org/10.1074/jbc.M806026200
Tojo H, Asano T, Kato K, Udaka S, Horiuchi R, Kakinuma A (1994) Production of human protein disulfide isomerase by Bacillus brevis. J Biotechnol 33:55–62. https://doi.org/10.1016/0168-1656(94)90098-1
Valentini E, Kikhney AG, Previtali G, Jeffries CM, Svergun DI (2015) SASBDB, a repository for biological small-angle scattering data. Nucleic Acids Res 43:D357–D363. https://doi.org/10.1093/nar/gku1047
Venetianer P, Straub FB (1963) The enzymic reactivation of reduced ribonuclease. Biochim Biophys Acta 67:166–168. https://doi.org/10.1016/0926-6569(63)90223-2
Volkov VV, Svergun DI (2003) Uniqueness of ab initio shape determination in small-angle scattering. J Appl Crystallogr. https://doi.org/10.1107/S0021889803000268
von Castelmur E, Strümpfer J, Franke B, Bogomolovas J, Barbieri S, Qadota H, Konarev PV, Svergun DI, Labeit S, Benian GM, Schulten K, Mayans O (2012) Identification of an N-terminal inhibitory extension as the primary mechanosensory regulator of twitchin kinase. Proc Natl Acad Sci U S A 109:13608–13613. https://doi.org/10.1073/pnas.1200697109
Walker KW, Gilbert HF (1997) Scanning and escape during protein-disulfide isomerase- assisted protein folding. J Biol Chem 272:8845–8848. https://doi.org/10.1074/jbc.272.14.8845
Wallis AK, Sidhu A, Byrne LJ, Howard MJ, Ruddock LW, Williamson RA, Freedman RB (2009) The ligand-binding b’ domain of human protein disulphide-isomerase mediates homodimerization. Protein Sci 18:2569–2577. https://doi.org/10.1002/pro.270
Wang C, Chen S, Wang X, Wang L, Wallis AK, Freedman RB, Wang C (2010) Plasticity of human protein disulfide isomerase: evidence for mobility around the X-linker region and its functional significance. J Biol Chem 285:26788–26797. https://doi.org/10.1074/jbc.M110.107839
Wang C, Li W, Ren J, Fang J, Ke H, Gong W, Feng W, Wang C-C (2013) Structural insights into the redox-regulated dynamic conformations of human protein disulfide isomerase. Antioxid Redox Signal 19:36–45. https://doi.org/10.1089/ars.2012.4630
Wang L, Wang X, Wang C (2015) Protein disulfide-isomerase, a folding catalyst and a redox- regulated chaperone. Free Radic Biol Med 83:305–313. https://doi.org/10.1016/j.freeradbiomed.2015.02.007
Weissman JS, Kim PS (1993) Efficient catalysis of disulphide bond rearrangements by protein disulphide isomerase. Nature 365:185–188. https://doi.org/10.1038/365185a0
Wetterau JR, Combs KA, Spinner SN, Joiner BJ (1990) Protein disulfide isomerase is a component of the microsomal triglyceride transfer protein complex. J Biol Chem 265:9800–9807
Xiao R, Wilkinson B, Solovyov A, Winther JR, Holmgren A, Lundström-Ljung J, Gilbert HF (2004) The contributions of protein disulfide isomerase and its homologues to oxidative protein folding in the yeast endoplasmic reticulum. J Biol Chem 279:49780–49786. https://doi.org/10.1074/jbc.M409210200
Yang IS, Kim TG, Park BS, Cho KJ, Lee J-H, Park Y, Kim KH (2010) Crystal structures of aprotinin and its complex with sucrose octasulfate reveal multiple modes of interactions with implications for heparin binding. Biochem Biophys Res Commun 397:429–435. https://doi.org/10.1016/j.bbrc.2010.05.113
Yu XC, Wang CC, Tsou CL (1994) Association and dissociation of protein disulfide isomerase. Biochim Biophys Acta 1207:109–113. https://doi.org/10.1016/0167-4838(94)90058-2
Acknowledgements
S.M. and C.S. thank the BioCAT advanced SAXS training course for the static measurement. We also thank Dr. Jesse Hopkins for his assistance in technical details about the instrument, Dr. Srinivas Chakravarthy, and APS 18ID/ BioCAT Beamline for SAXS data collection. This research used resources from the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. "This project was supported by grant 9 P41 GM103622 from the National Institute of General Medical Sciences of the National Institutes of Health". The use of the Pilatus 3 1M detector was provided by grant 1S10OD018090-01 from NIGMS. The content is solely the responsibility of the authors and does not necessarily reflect the official views of the National Institute of General Medical Sciences or the National Institutes of Health. Molecular graphics and analyses were performed with UCSF Chimera, developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from NIH P41-GM103311.
Funding
This work was supported by the grants from the Department of Biotechnology (DBT) NO.BT/PR13689/BRB/10/820/2010 and University Grants Commission (UGC)-Raman fellowship F.NO.5-1/2013(IC), Government of India.
Author information
Authors and Affiliations
Contributions
C.S. and S.B. Designed the research, performed the research, analyzed the data, and wrote the manuscript. T.C. Provided resources and technical assistance. N.V., P.K., and R.B. Provided Technical assistance and suggestions to refine this research. S.M. Designed the research, provided the resources, acquired funds.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
40203_2024_198_MOESM1_ESM.docx
Appendix A. Supplementary data. The Supplementary material is available as a separate document which includes the DEAE elution profile, enzyme activity assay results, Size Exclusion elution profile, and SDS-PAGE analysis of PDI purification. Statistics of SAXS analysis, scattering and CRYSOL Goodness of fit Plot for all the protein samples, Comparison of BPTI, and RNase solution structure with the existing crystal structure, SASBDB Accession Codes for the structures determined. Supplementary file1 (DOCX 1511 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sanyasi, C., Balakrishnan, S.S., Chinnasamy, T. et al. Insights on the dynamic behavior of protein disulfide isomerase in the solution environment through the SAXS technique. In Silico Pharmacol. 12, 23 (2024). https://doi.org/10.1007/s40203-024-00198-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s40203-024-00198-0