Abstract
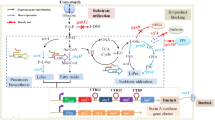
A novel xylanase gene (denominated xynDRTY1) was identified from Tengchong hot spring by a metagenomic approach. Its amino acid sequence was 73.43% identical to a hypothetical protein from Bryobacterales bacterium. The codon-optimized XynDRTY1 gene was synthesized and overexpressed in Escherichia coli. The XynDRTY1 was purified by using Ni–NTA affinity chromatography. It exhibited activity with natural glycosides, such as beechwood xylan (21.2 ± 3 U/mg) and oat spelt xylan (8.2 ± 0.3 U/mg). Its optimum pH was determined to be 6.0 and optimum temperature of 65 ℃, along with its stability over 140% and 110% relative enzyme activity after incubation at 60 ℃ for 20 min and 120 min, respectively. Based on these findings, we believe that XynDRTY1, as thermostable xylanase, may prove useful for biotechnological applications.




Similar content being viewed by others
References
Ghadikolaei KK, Sangachini ED, Vahdatirad V, Noghabi KA, Zahiri HS (2019) An extreme halophilic xylanase from camel rumen metagenome with elevated catalytic activity in high salt concentrations. AMB Express 9(1):86. https://doi.org/10.1186/s13568-019-0809-2
Takkellapati S, Li T, Gonzalez MA (2018) An overview of biorefinery-derived platform chemicals from a cellulose and hemicellulose biorefinery. Clean Technol Environ Policy 20(7):1615–1630. https://doi.org/10.1007/s10098-018-1568-5
Samanta AK, Jayapal N, Kolte AP, Senani S, Sridhar M, Dhali A, Suresh KP, Jayaram C, Prasad CS (2014) Process for enzymatic production of xylooligosaccharides from the xylan of corn cobs. J Food Process Preserv 39(6):729–736. https://doi.org/10.1111/jfpp.12282
Tenkanen M, Puls J, Poutanen K (1992) Two major xylanases of Trichoderma reesei. Enzyme Microb Technol 14(7):566–574. https://doi.org/10.1016/0141-0229(92)90128-b
López-López O, Cerdán M, González-Siso M (2013) Hot spring metagenomics. Life 3(2):308–320. https://doi.org/10.3390/life3020308
Knapik K, Becerra M, González-Siso M-I (2019) Microbial diversity analysis and screening for novel xylanase enzymes from the sediment of the lobios hot spring in Spain. Sci Rep 9(1):11195. https://doi.org/10.1038/s41598-019-47637-z
Kurazono H, Pal A, Bag PK, Balakrish Nair G, Karasawa T, Mihara T, Takeda Y (1995) Distribution of genes encoding cholera toxin, zonula occludens toxin, accessory cholera toxin, and El Tor hemolysin Vibrio cholerae of diverse origins. Microb Pathog 18(3):231–235. https://doi.org/10.1016/s0882-4010(95)90076-4
Uchiyama T, Miyazaki K (2009) Functional metagenomics for enzyme discovery: challenges to efficient screening. Curr Opin Biotechnol 20(6):616–622. https://doi.org/10.1016/j.copbio.2009.09.010
Tiwari R, Nain L, Labrou NE, Shukla P (2017) Bioprospecting of functional cellulases from metagenome for second generation biofuel production: a review. Crit Rev Microbiol 44(2):244–257. https://doi.org/10.1080/1040841x.2017.1337713
Zerbino DR, Birney E (2008) Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18(5):821–829. https://doi.org/10.1101/gr.074492.107
Tatusov RL (2001) The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res 29(1):22–28. https://doi.org/10.1093/nar/29.1.22
Nakaya A, Katayama T, Itoh M, Hiranuka K, Kawashima S, Moriya Y, Goto S (2012) KEGG OC: a large-scale automatic construction of taxonomy-based ortholog clusters. Nucleic Acids Res 41(D1):D353–D357. https://doi.org/10.1093/nar/gks1239
Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Bateman A (2007) The Pfam protein families database. Nucleic Acids Res 36(Database), D281–D288. https://doi.org/10.1093/nar/gkm960
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882. https://doi.org/10.1093/nar/25.24.4876
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874. https://doi.org/10.1093/molbev/msw054
Šali A, Blundell TL (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234(3):779–815. https://doi.org/10.1006/jmbi.1993.1626
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23(21):2947–2948. https://doi.org/10.1093/bioinformatics/btm404
Gouet P, Robert X, Courcelle E (2003) ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res 31(13):3320–3323. https://doi.org/10.1007/s10404-008-0309-1
Yin YR, Meng ZH, Hu QW, Jiang Z, Xian WD, Li LH, Li WJ (2017) The Hybrid Strategy of Thermoactinospora rubra YIM 77501T for utilizing cellulose as a carbon source at different temperatures. Front Microbiol 8:942. https://doi.org/10.3389/fmicb.2017.00942
Yin YR, Hu QW, Xian WD, Feng Z, Zhou EM, Hong M, Min X, Zhi XY, Li WJ (2016) Characterization of a neutral recombinant xylanase from Thermoactinospora rubra YIM 77501T. Antonie Van Leeuwenhoek 110:429–436. https://doi.org/10.1007/s10482-016-0798-y
Miller GL (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31(3):426–428. https://doi.org/10.1021/ac60147a030
Lo Leggio L, Kalogiannis S, Bhat MK, Pickersgill RW (1999) High resolution structure and sequence of T. aurantiacus xylanase I: implications for the evolution of thermostability in family 10 xylanases and enzymes with (beta)alpha-barrel architecture. Proteins 36:295–306. https://doi.org/10.1002/(sici)1097-0134(19990815)36:3%3c295::aid-prot4%3e3.0.co;2-6
Niderhaus C, Garrido M, Insani M, Campos E, Wirth S (2018) Heterologous production and characterization of a thermostable GH10 family endo-xylanase from Pycnoporus sanguineus bafc 2126. Process Biochem 67(APR.):92–98. https://doi.org/10.1016/j.procbio.2018.01.017
Bhalla A, Bansal N, Kumar S, Bischoff KM, Sani RK (2013) Improved lignocellulose conversion to biofuels with thermophilic bacteria and thermostable enzymes. Bioresource Technol 128(Complete):751–759. https://doi.org/10.1016/j.biortech.2012.10.145
He J, Tang F, Chen D, Yu B, Luo Y, Zheng P, Mao X, Yu J, Yu F (2019) Design, expression and functional characterization of a thermostable xylanase from Trichoderma reesei. PLoS One 14(1):e0210548. https://doi.org/10.1371/journal.pone.0210548
Bai W, Xue Y, Zhou C, Ma Y (2012) Cloning, expression and characterization of a novel salt-tolerant xylanase from Bacillus sp. SN5. Biotechnol Lett 34(11):2093–2099. https://doi.org/10.1007/s10529-012-1011-7
Jacomini D, Bussler L, Corrêa JM, Kadowaki MK, Simo R (2020) Cloning, expression and characterization of C. crescentus xyna2 gene and application of xylanase II in the deconstruction of plant biomass. Mol Biol Rep 47(6):4427–4438. https://doi.org/10.1007/s11033-020-05507-2
Loaces I, Bottini G, Moyna G, Fabiano E, Martínez A, Noya F (2016) Endog: a novel multifunctional halotolerant glucanase and xylanase isolated from cow rumen. J Mol Catal B Enzym S1381117716300042.https://doi.org/10.1016/j.molcatb.2016.01.004
Chapla D, Pandit P, Shah A (2012) Production of xylooligosaccharides from corncob xylan by fungal xylanase and their utilization by probiotics. Bioresource Technol 115(none):215–221. https://doi.org/10.1016/j.biortech.2011.10.083
Finegold SM, Li Z, Summanen PH, Downes J, Thames G, Corbett K et al (2014) Xylooligosaccharide increases bifidobacteria but not lactobacilli in human gut microbiota. Food Funct 5(3):436. https://doi.org/10.1039/c3fo60348b
Chen M, Liu S, Imam K, Sun L, Wang Y, Gu T et al (2020) The effect of xylooligosaccharide, xylan, and whole wheat bran on the human gut bacteria. Front Microbiol 11:2936–2947. https://doi.org/10.3389/fmicb.2020.568457
Acknowledgements
We gratefully acknowledge Prof. Wen-Jun Li and his team from Sun Yat-sen University for their guidance on our metagenomic data compilation and analysis.
Funding
This research was supported by the National Natural Sciences Foundation of China Regional Program (Grant Nos. 31660015 and 31860243) and Yunnan Applied Basic Research Projects (Grant Nos. 202101AU070138, 2017FB024, and 2017FH001-032).
Author information
Authors and Affiliations
Contributions
YRY, PS, and LQY conceived the study. YRY and WH cloned the gene and cultured strains. RFY and HYL purified the recombinant protein. LL measured enzymatic activity. XWL and ZLL performed the data analysis and mapping. YRY, WH, PS, and LQY wrote the manuscript. All authors discussed the results and commented on the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval
This article does not contain any studies related to human participants or animals.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yin, YR., Li, L., Yang, RF. et al. Characterization of a metagenome-derived thermostable xylanase from Tengchong hot spring. Biomass Conv. Bioref. 14, 10027–10034 (2024). https://doi.org/10.1007/s13399-022-03296-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13399-022-03296-1