Abstract
Relevance
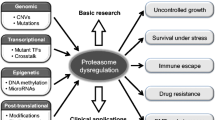
Proteasome, a cylindrical complex containing 19S regulatory particle lid, 19S regulatory particle base, and 20S core particle, acted as a major mechanism to regulate the levels of intracellular proteins and degrade misfolded proteins, which involved in many cellular processes, and played important roles in cancer biological processes. Elucidation of proteasome alterations across multiple cancer types will directly contribute to cancer medical services in the context of predictive, preventive, and personalized medicine (PPPM / 3P medicine).
Purpose
This study aimed to investigate proteasome gene alterations across 33 cancer types for discovery of effective biomarkers and therapeutic targets in the framework of PPPM practice in cancers.
Methods
Proteasome gene data, including gene expression RNAseq, somatic mutation, tumor mutation burden (TMB), copy number variant (CNV), microsatellite instability (MSI) score, clinical characteristics, immune phenotype, 22 immune cells, cancer stemness index, drug sensitivity, and related pathways, were systematically analyzed with publically available database and bioinformatics across 11,057 patients with 33 cancer types.
Results
Differentially expressed proteasome genes were extensively found between tumor and control tissues. PSMB4 occurred the top mutation event among proteasome genes, and those proteasome genes were significantly associated with TMB and MSI score. Most of proteasome genes were positively related to CNV among single deletion, control copy number, and single gain. Kaplan–Meier curves and COX regression survival analysis showed proteasome genes were significantly associated with patient survival rate across 33 cancer types. Furthermore, the expressions of proteasome genes were significantly different among different clinical stages and immune subtypes. The expressions of proteasome genes were correlated with immune-related scores (ImmuneScore, StromalScore, and ESTIMATEScore), 22 immune cells, and cancer stemness. The sensitivities of multiple drugs were closely related to proteasome gene expressions. The identified proteasome and proteasome-interacted proteins were significantly enriched in various cancer-related pathways.
Conclusions
This study provided the first landscape of proteasome alterations across 11,057 patients with 33 cancer types and revealed that proteasome played a significant and wide functional role in cancer biological processes. These findings are the precious scientific data to reveal the common and specific alterations of proteasome genes among 33 cancer types, which benefits the research and practice of PPPM in cancers.








Similar content being viewed by others
Data accessibility
All data and materials were provided in this manuscript and supplementary materials.
Abbreviations
- ACC:
-
Adrenocortical carcinoma
- ADRM1:
-
Proteasome 26S subunit, non-ATPase 13
- BLCA:
-
Bladder urothelial carcinoma
- BP:
-
Biological process
- BRCA:
-
Breast cancer
- CC:
-
Cellular component
- CDKs:
-
Cyclin-dependent kinases
- CESC:
-
Cervical squamous cell carcinoma and endocervical adenocarcinoma
- CHOL:
-
Cholangiocarcinoma
- CNV:
-
Copy number variant
- COAD:
-
Colon adenocarcinoma
- DEGs:
-
Differentially expressed genes
- DLBC:
-
Lymphoid neoplasm diffuse large b-cell lymphoma
- DTP:
-
Developmental therapeutics program
- ESCA:
-
Esophageal carcinoma
- GBM:
-
Glioblastoma multiforme
- GO:
-
Gene Ontology
- GSVA:
-
Gene set variation analysis
- HNSC:
-
Head and neck squamous carcinoma
- HR:
-
Hazard ratio
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- KICH:
-
Kidney chromophobe
- KIRC:
-
Kidney renal clear cell carcinoma
- KIRP:
-
Kidney renal papillary cell carcinoma
- LAML:
-
Acute myeloid leukemia
- LGG:
-
Brain lower grade glioma
- LIHC:
-
Liver hepatocellular carcinoma
- LUAD:
-
Lung adenocarcinoma
- LUSC:
-
Lung squamous cell carcinoma
- MESO:
-
Mesothelioma
- MF:
-
Molecular function
- MHC:
-
Major histocompatibility complex class I
- MMR:
-
DNA mismatch repair
- mRNA:
-
Messenger RNA
- MSI:
-
Microsatellite instability
- OV:
-
Ovarian serous cystadenocarcinoma
- PAAD:
-
Pancreatic adenocarcinoma
- PCPG:
-
Pheochromocytoma and paraganglioma
- PRAD:
-
Prostate adenocarcinoma
- PSMA1:
-
Proteasome 20S subunit alpha 1
- PSMA2:
-
Proteasome 20S subunit alpha 2
- PSMA3:
-
Proteasome 20S subunit alpha 3
- PSMA4:
-
Proteasome 20S subunit alpha 4
- PSMA5:
-
Proteasome 20S subunit alpha 5
- PSMA6:
-
Proteasome 20S subunit alpha 6
- PSMA7:
-
Proteasome 20S subunit alpha 7
- PSMB1:
-
Proteasome 20S subunit beta 1
- PSMB10:
-
Proteasome 20S subunit beta 10
- PSMB11:
-
Proteasome subunit beta 11
- PSMB2:
-
Proteasome 20S subunit beta 2
- PSMB3:
-
Proteasome 20S subunit beta 3
- PSMB4:
-
Proteasome 20S subunit beta 4
- PSMB5:
-
Proteasome 20S subunit beta 5
- PSMB6:
-
Proteasome 20S subunit beta 6
- PSMB7:
-
Proteasome 20S subunit beta 7
- PSMB8:
-
Proteasome 20S subunit beta 8
- PSMB9:
-
Proteasome 20S subunit beta 9
- PSMC1:
-
Proteasome 26S subunit, ATPase 1
- PSMC2:
-
Proteasome 26S subunit, ATPase 2
- PSMC3:
-
Proteasome 26S subunit, ATPase 3
- PSMC4:
-
Proteasome 26S subunit, ATPase 4
- PSMC5:
-
Proteasome 26S subunit, ATPase 5
- PSMC6:
-
Proteasome 26S subunit, ATPase 6
- PSMD1:
-
Proteasome 26S subunit, non-ATPase 1
- PSMD11:
-
Proteasome 26S subunit, non-ATPase 11
- PSMD12:
-
Proteasome 26S subunit, non-ATPase 12
- PSMD13:
-
Proteasome 26S subunit, non-ATPase 13
- PSMD14:
-
Proteasome 26S subunit, non-ATPase 14
- PSMD2:
-
Proteasome 26S subunit, non-ATPase 2
- PSMD3:
-
Proteasome 26S subunit, non-ATPase 3
- PSMD4:
-
Proteasome 26S subunit, non-ATPase 4
- PSMD6:
-
Proteasome 26S subunit, non-ATPase 6
- PSMD7:
-
Proteasome 26S subunit, non-ATPase 7
- PSMD8:
-
Proteasome 26S subunit, non-ATPase 8
- PSMD9:
-
Proteasome 26S subunit, non-ATPase 9
- PSME1:
-
Proteasome activator subunit 1
- PSME2:
-
Proteasome activator subunit 2
- PSME3:
-
Proteasome activator subunit 3
- PSME4:
-
Proteasome activator subunit 4
- READ:
-
Rectum adenocarcinoma
- RNAss:
-
RNA expression-based stemness scores
- SARC:
-
Sarcoma
- SEM1:
-
SEM1 26S proteasome complex subunit
- SKCM:
-
Skin cutaneous melanoma
- STAD:
-
Stomach adenocarcinoma
- TCGA:
-
The Cancer Genome Atlas
- TGCT:
-
Testicular germ cell tumors
- THCA:
-
Thyroid carcinoma
- THYM:
-
Thymoma
- TMB:
-
Tumor mutation burden
- TME:
-
Tumor microenvironment
- UCEC:
-
Uterine corpus endometrial carcinoma
- UCS:
-
Uterine carcinosarcoma
- UVM:
-
Uveal melanoma
References
Spits M, Janssen LJ, Voortman LM, Kooij R, Neefjes ACM, Ovaa H, et al. Homeostasis of soluble proteins and the proteasome post nuclear envelope reformation in mitosis. J Cell Sci. 2019; 132. https://doi.org/10.1242/jcs.225524.
Limanaqi F, Biagioni F, Gambardella S, Familiari P, Frati A, Fornai F. Promiscuous roles of autophagy and proteasome in neurodegenerative proteinopathies. Int J Mol Sci. 2020; 21. https://doi.org/10.3390/ijms21083028.
Bard JAM, Goodall EA, Greene ER, Jonsson E, Dong KC, Martin A. Structure and function of the 26S proteasome. Annu Rev Biochem. 2018;87:697–724. https://doi.org/10.1146/annurev-biochem-062917-011931.
Gierisch ME, Giovannucci TA, Dantuma NP. Reporter-based screens for the ubiquitin/proteasome system. Front Chem. 2020;8:64. https://doi.org/10.3389/fchem.2020.00064.
Budenholzer L, Cheng CL, Li Y, Hochstrasser M. Proteasome structure and assembly. J Mol Biol. 2017;429:3500–24. https://doi.org/10.1016/j.jmb.2017.05.027.
Coux O, Zieba BA, Meiners S. The proteasome system in health and disease. Adv Exp Med Biol. 2020;1233:55–100. https://doi.org/10.1007/978-3-030-38266-7_3.
Harrigan JA, Jacq X, Martin NM, Jackson SP. Deubiquitylating enzymes and drug discovery: emerging opportunities. Nat Rev Drug Discov. 2018;17:57–78. https://doi.org/10.1038/nrd.2017.152.
Groll M, Huber R. Purification, crystallization, and X-ray analysis of the yeast 20S proteasome. Methods Enzymol. 2005;398:329–36. https://doi.org/10.1016/s0076-6879(05)98027-0.
Zhang S,Mao Y. AAA+ ATPases in protein degradation: structures, functions and mechanisms. Biomolecules. 2020; 10. https://doi.org/10.3390/biom10040629.
Thibaudeau TA, Smith DM. A practical review of proteasome pharmacology. Pharmacol Rev. 2019;71:170–97. https://doi.org/10.1124/pr.117.015370.
Rousseau A, Bertolotti A. Regulation of proteasome assembly and activity in health and disease. Nat Rev Mol Cell Biol. 2018;19:697–712. https://doi.org/10.1038/s41580-018-0040-z.
Sharma A, Trivedi AK. Regulation of apoptosis by E3 ubiquitin ligases in ubiquitin proteasome system. Cell Biol Int. 2020;44:721–34. https://doi.org/10.1002/cbin.11277.
Neutzner A, Li S, Xu S, Karbowski M. The ubiquitin/proteasome system-dependent control of mitochondrial steps in apoptosis. Semin Cell Dev Biol. 2012;23:499–508. https://doi.org/10.1016/j.semcdb.2012.03.019.
Wójcik C, DeMartino GN. Intracellular localization of proteasomes. Int J Biochem Cell Biol. 2003;35:579–89. https://doi.org/10.1016/s1357-2725(02)00380-1.
Frezza M, Schmitt S, Dou QP. Targeting the ubiquitin-proteasome pathway: an emerging concept in cancer therapy. Curr Top Med Chem. 2011;11:2888–905. https://doi.org/10.2174/156802611798281311.
Drexler HC. The role of p27Kip1 in proteasome inhibitor induced apoptosis. Cell Cycle. 2003;2:438–41.
Bonet-Costa V, Pomatto LC, Davies KJ. The proteasome and oxidative stress in Alzheimer’s disease. Antioxid Redox Signal. 2016;25:886–901. https://doi.org/10.1089/ars.2016.6802.
Davies MJ. Protein oxidation and peroxidation. Biochem J. 2016;473:805–25. https://doi.org/10.1042/bj20151227.
Kammerl IE, Meiners S. Proteasome function shapes innate and adaptive immune responses. Am J Physiol Lung Cell Mol Physiol. 2016;311:L328–36. https://doi.org/10.1152/ajplung.00156.2016.
Kloetzel PM, Ossendorp F. Proteasome and peptidase function in MHC-class-I-mediated antigen presentation. Curr Opin Immunol. 2004;16:76–81. https://doi.org/10.1016/j.coi.2003.11.004.
Murata S, Takahama Y, Kasahara M, Tanaka K. The immunoproteasome and thymoproteasome: functions, evolution and human disease. Nat Immunol. 2018;19:923–31. https://doi.org/10.1038/s41590-018-0186-z.
Dragnev KH, Freemantle SJ, Spinella MJ, Dmitrovsky E. Cyclin proteolysis as a retinoid cancer prevention mechanism. Ann N Y Acad Sci. 2001;952:13–22. https://doi.org/10.1111/j.1749-6632.2001.tb02724.x.
Voutsadakis IA. Proteasome expression and activity in cancer and cancer stem cells. Tumour Biol. 2017;39:1010428317692248. https://doi.org/10.1177/1010428317692248.
Lu M, Zhan X. The crucial role of multiomic approach in cancer research and clinically relevant outcomes. EPMA J. 2018;9:77–102. https://doi.org/10.1007/s13167-018-0128-8.
Berryman K, Buhimschi CS, Zhao G, Axe M, Locke M, Buhimschi IA. Proteasome levels and activity in pregnancies complicated by severe preeclampsia and hemolysis, elevated liver enzymes, and thrombocytopenia (HELLP) syndrome. Hypertension. 2019;73:1308–18. https://doi.org/10.1161/hypertensionaha.118.12437.
Cheng T, Zhan X. Pattern recognition for predictive, preventive, and personalized medicine in cancer. EPMA J. 2017;8:51–60. https://doi.org/10.1007/s13167-017-0083-9.
Hu R, Wang X, Zhan X. Multi-parameter systematic strategies for predictive, preventive and personalised medicine in cancer. EPMA J. 2013;4:2. https://doi.org/10.1186/1878-5085-4-2.
Yamamoto H, Imai K. Microsatellite instability: an update. Arch Toxicol. 2015;89:899–921. https://doi.org/10.1007/s00204-015-1474-0.
Mofers A, Pellegrini P, Linder S, D’Arcy P. Proteasome-associated deubiquitinases and cancer. Cancer Metastasis Rev. 2017;36:635–53. https://doi.org/10.1007/s10555-017-9697-6.
Catalgol B. Proteasome and cancer. Prog Mol Biol Transl Sci. 2012;109:277–93. https://doi.org/10.1016/b978-0-12-397863-9.00008-0.
Chen Y, Zhang Y, Guo X. Proteasome dysregulation in human cancer: implications for clinical therapies. Cancer Metastasis Rev. 2017;36:703–16. https://doi.org/10.1007/s10555-017-9704-y.
Ivanova EV, Cheremisina OV, Afanasiev SG, Kondakova IV. Proteasome and calpain activities in colon cancer: the relation with metastasis and prognosis. Vopr Onkol. 2016;62:794–800.
Kondakova IV, Spirina LV, Shashova EE, Koval VD, Kolomiets LA, Chernysheva AL, et al. Proteasome activity in tumors of female reproductive system. Bioorg Khim. 2012;38:106–10. https://doi.org/10.1134/s106816201201013x.
Rohondia SO, Ahmed ZSO, Dou QP. Updated review and perspective on 20S proteasome inhibitors in the treatment of lung cancer. Curr Cancer Drug Targets. 2020;20:392–409. https://doi.org/10.2174/1568009620666200226094000.
Raninga PV, Lee A, Sinha D, Dong LF, Datta KK, Lu X, et al. Marizomib suppresses triple-negative breast cancer via proteasome and oxidative phosphorylation inhibition. Theranostics. 2020;10:5259–75. https://doi.org/10.7150/thno.42705.
Köster F, Sauer L, Hoellen F, Ribbat-Idel J, Bräutigam K, Rody A, et al. PSMD9 expression correlates with recurrence after radiotherapy in patients with cervical cancer. Oncol Lett. 2020;20:581–8. https://doi.org/10.3892/ol.2020.11622.
Wu W, Zhong J, Chen J, Niu P, Ding Y, Han S, et al. Prognostic and therapeutic significance of adhesion-regulating molecule 1 in estrogen receptor-positive breast cancer. Clin Breast Cancer. 2020;20:131-144.e3. https://doi.org/10.1016/j.clbc.2019.07.009.
Li J, Zou C, Bai Y, Wazer DE, Band V, Gao Q. DSS1 is required for the stability of BRCA2. Oncogene. 2006;25:1186–94. https://doi.org/10.1038/sj.onc.1209153.
Bista R, Lee DW, Pepper OB, Azorsa DO, Arceci RJ, Aleem E. Disulfiram overcomes bortezomib and cytarabine resistance in Down-syndrome-associated acute myeloid leukemia cells. J Exp Clin Cancer Res. 2017;36:22. https://doi.org/10.1186/s13046-017-0493-5.
Zhang Z, Clawson A, Realini C, Jensen CC, Knowlton JR, Hill CP, et al. Identification of an activation region in the proteasome activator REGalpha. Proc Natl Acad Sci U S A. 1998;95:2807–11. https://doi.org/10.1073/pnas.95.6.2807.
Tafe LJ. Non-small cell lung cancer as a precision oncology paradigm: emerging targets and tumor mutational burden (TMB). Adv Anat Pathol. 2020;27:3–10. https://doi.org/10.1097/pap.0000000000000244.
Lee GY, Haverty PM, Li L, Kljavin NM, Bourgon R, Lee J, et al. Comparative oncogenomics identifies PSMB4 and SHMT2 as potential cancer driver genes. Cancer Res. 2014;74:3114–26. https://doi.org/10.1158/0008-5472.Can-13-2683.
Song L, Ma N, Han L, Yan H, Yan B, Yuan Z, et al. Association between LMP2/LMP7 genetic variability and the metastasis risk of ovarian cancer in Chinese women in Beijing. Hum Immunol. 2014;75:239–44. https://doi.org/10.1016/j.humimm.2013.12.006.
Rouette A, Trofimov A, Haberl D, Boucher G, Lavallée VP, D’Angelo G, et al. Expression of immunoproteasome genes is regulated by cell-intrinsic and -extrinsic factors in human cancers. Sci Rep. 2016;6:34019. https://doi.org/10.1038/srep34019.
Lee M, Song IH, Heo SH, Kim YA, Park IA, Bang WS, et al. Expression of immunoproteasome subunit LMP7 in breast cancer and its association with immune-related markers. Cancer Res Treat. 2019;51:80–9. https://doi.org/10.4143/crt.2017.500.
Li C, Dai S, Yan Z, Zhang X, Liu S, Wang X, et al. Genetic polymorphisms of proteasome subunit genes of the MHC-I antigen-presenting system are associated with cervical cancer in a Chinese Han population. Hum Immunol. 2020. https://doi.org/10.1016/j.humimm.2020.07.002.
Kalaora S, Lee JS, Barnea E, Levy R, Greenberg P, Alon M, et al. Immunoproteasome expression is associated with better prognosis and response to checkpoint therapies in melanoma. Nat Commun. 2020;11:896. https://doi.org/10.1038/s41467-020-14639-9.
Yi Z, Yang D, Liao X, Guo F, Wang Y, Wang X. PSME3 induces epithelial-mesenchymal transition with inducing the expression of CSC markers and immunosuppression in breast cancer. Exp Cell Res. 2017;358:87–93. https://doi.org/10.1016/j.yexcr.2017.05.017.
Seo D, Jung SM, Park JS, Lee J, Ha J, Kim M, et al. The deubiquitinating enzyme PSMD14 facilitates tumor growth and chemoresistance through stabilizing the ALK2 receptor in the initiation of BMP6 signaling pathway. EBioMedicine. 2019;49:55–71. https://doi.org/10.1016/j.ebiom.2019.10.039.
Cokic VP, Beleslin-Cokic BB, Noguchi CT, Schechter AN. Hydroxyurea increases eNOS protein levels through inhibition of proteasome activity. Nitric Oxide. 2007;16:371–8. https://doi.org/10.1016/j.niox.2007.01.001.
Takahashi K, Inukai T, Imamura T, Yano M, Tomoyasu C, Lucas DM, et al. Anti-leukemic activity of bortezomib and carfilzomib on B-cell precursor ALL cell lines. PLoS ONE. 2017;12: e0188680. https://doi.org/10.1371/journal.pone.0188680.
Ogura M. Current treatment strategy in mantle cell lymphoma. Nihon Rinsho. 2014;72:499–511.
Ji CH, Kwon YT. Crosstalk and interplay between the ubiquitin-proteasome system and autophagy. Mol Cells. 2017;40:441–9. https://doi.org/10.14348/molcells.2017.0115.
Qi SM, Cheng G, Cheng XD, Xu Z, Xu B, Zhang WD, et al. Targeting USP7-mediated deubiquitination of MDM2/MDMX-p53 pathway for cancer therapy: are we there yet? Front Cell Dev Biol. 2020;8:233. https://doi.org/10.3389/fcell.2020.00233.
Maliński M, Cichocki M. Proteasome inhibition as a new strategy in cancer therapy and chemoprevention. Postepy Hig Med Dosw (Online). 2013;67:90–106. https://doi.org/10.5604/17322693.1035963.
Bitzer A, Basler M, Groettrup M. Chaperone BAG6 is dispensable for MHC class I antigen processing and presentation. Mol Immunol. 2016;69:99–105. https://doi.org/10.1016/j.molimm.2015.11.004.
Cascio P, Hilton C, Kisselev AF, Rock KL, Goldberg AL. 26S proteasomes and immunoproteasomes produce mainly N-extended versions of an antigenic peptide. EMBO J. 2001;20:2357–66. https://doi.org/10.1093/emboj/20.10.2357.
Arata Y, Watanabe A, Motosugi R, Murakami R, Goto T, Hori S, et al. Defective induction of the proteasome associated with T-cell receptor signaling underlies T-cell senescence. Genes Cells. 2019;24:801–13. https://doi.org/10.1111/gtc.12728.
Jazirehi AR, Economou JS. Proteasome inhibition blocks NF-κB and ERK1/2 pathways, restores antigen expression, and sensitizes resistant human melanoma to TCR-engineered CTLs. Mol Cancer Ther. 2012;11:1332–41. https://doi.org/10.1158/1535-7163.Mct-11-0814.
Baloghova N, Lidak T, Cermak L. Ubiquitin ligases involved in the regulation of Wnt, TGF-β, and Notch signaling pathways and their roles in mouse development and homeostasis. Genes (Basel). 2019; 10. https://doi.org/10.3390/genes10100815.
Anastas JN, Moon RT. WNT signalling pathways as therapeutic targets in cancer. Nat Rev Cancer. 2013;13:11–26. https://doi.org/10.1038/nrc3419.
Fabre B, Livneh I, Ziv T, Ciechanover A. Identification of proteins regulated by the proteasome following induction of endoplasmic reticulum stress. Biochem Biophys Res Commun. 2019;517:188–92. https://doi.org/10.1016/j.bbrc.2019.07.040.
Ji L, Lu B, Zamponi R, Charlat O, Aversa R, Yang Z, et al. USP7 inhibits Wnt/β-catenin signaling through promoting stabilization of Axin. Nat Commun. 2019;10:4184. https://doi.org/10.1038/s41467-019-12143-3.
Espinoza I, Miele L. Notch inhibitors for cancer treatment. Pharmacol Ther. 2013;139:95–110. https://doi.org/10.1016/j.pharmthera.2013.02.003.
Yeh CH, Bellon M, Nicot C. FBXW7: a critical tumor suppressor of human cancers. Mol Cancer. 2018;17:115. https://doi.org/10.1186/s12943-018-0857-2.
Zhang F, Laiho M. On and off: proteasome and TGF-beta signaling. Exp Cell Res. 2003;291:275–81. https://doi.org/10.1016/j.yexcr.2003.07.007.
Wang T. The 26S proteasome system in the signaling pathways of TGF-beta superfamily. Front Biosci. 2003;8:d1109–27. https://doi.org/10.2741/1057.
Golubnitschaja O, Liskova A, Koklesova L, Samec M, Biringer K, Büsselberg D, Podbielska H, Kunin AA, Evsevyeva ME, Shapira N, Paul F, Erb C, Dietrich DE, Felbel D, Karabatsiakis A, Bubnov R, Polivka J, Polivka J Jr, Birkenbihl C, Fröhlich H, Hofmann-Apitius M, Kubatka P. Caution, “normal” BMI: health risks associated with potentially masked individual underweight—EPMA Position Paper 2021. EPMA J. 2021;17:1–22. https://doi.org/10.1007/s13167-021-00251-4.
Acknowledgements
We would like to thank The Cancer Genome Atlas (TCGA) project organizers as well as all study participants for providing the publically available TCGA RNA-seq data and clinical data.
Funding
This work was supported by the Shandong First Medical University Talent Introduction Funds (to X.Z.), the Hunan Provincial Hundred Talent Plan (to X.Z.), the National Natural Science Foundation of China (82172866), and the Academic Promotion Program of Shandong First Medical University (2019ZL002).
Author information
Authors and Affiliations
Contributions
N.L. designed, analyzed data, prepared figures and tables, and wrote the manuscript. X.Z. conceived the concept, supervised results, designed, wrote and critically revised manuscript, and was responsible for its financial supports and the corresponding works. All authors approved the final manuscript.
Corresponding author
Ethics declarations
Ethical approval
Not applicable.
Consent publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, N., Zhan, X. Integrated genomic analysis of proteasome alterations across 11,057 patients with 33 cancer types: clinically relevant outcomes in framework of 3P medicine. EPMA Journal 12, 605–627 (2021). https://doi.org/10.1007/s13167-021-00256-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13167-021-00256-z