Abstract
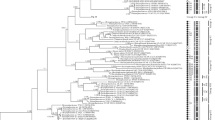
The correct identification of species diversity of small single-celled green coccoid microalgae still causes difficulties, since their relatively simple morphology hides a high physiological, ecological and genetic diversity. The use of molecular genetic methods has revolutionized the study of the true biodiversity of so-called “small green balls,” allowing the discovery of numerous new taxa. This article presents the results of a study of strains recently isolated from small freshwater urbanized lakes (Vasilievsky Lakes system, Samara region, Russian Federation). Morphologically, these strains were close to the genus Meyerella: spherical cells, cup-shaped, or wide girdle-shaped parietal chloroplast without pyrenoid. Analysis of the 18S–ITS1–5.8S–ITS2 sequences also showed that the studied strains belong to this genus. Comparison of morphological characteristics, habitat and lifestyle, analysis of tree topology, genetic distances and secondary structures of the ITS1 and ITS2 spacers of the Meyerella members, as well as the delimitation results using the Automatic Barcode Gap Discovery (ABGD) method, the Poisson Tree Processes (PTP) model, the Generalized Mixed Yule The Coalescent (GMYC) method allowed us to establish that the studied strains ACSSI 346, ACSSI 362 and ACSSI 363 are representatives of a new species – M. similis sp. nov.







Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Abbreviations
- ABGD:
-
Automatic Barcode Gap Discovery
- ACOI:
-
Coimbra Collection of Algae, Portugal
- ACSSI:
-
Algal Collection of the Soil Science Institute, Russia
- BI:
-
Bayesian inference
- BP:
-
Bootstrap proportion
- CBC:
-
Compensatory base change
- CCALA:
-
The Culture Collection of Autotrophic Organisms, The Czech Republic
- CCAP:
-
The Culture Centre Algae and Protozoa, UK
- CCMP:
-
The Culture Collection of Marine Phytoplankton, USA
- CV:
-
Coefficient of substitution rate variation
- GMYC:
-
Generalized mixed yule coalescent
- HPD:
-
Posterior density
- IPPAS:
-
Collection of microalgae and cyanobacteria IPPAS, Russia
- ITS1:
-
First internal transcribed spacer
- ITS2:
-
Second internal transcribed spacer
- LM:
-
Light microscopy
- ML:
-
Maximum likelihood
- MOTU:
-
Molecular operational taxonomic units
- NIES:
-
Microbial Culture Collection at the National Institute for Environmental Studies, Japan
- PP:
-
Posterior probability
- PCR:
-
Polymerase chain reaction
- PTP:
-
Poisson tree processes
- SAG:
-
Sammlung von Algenkulturen at the University of Göttingen, Germany
- TEM:
-
Transmission electron microscopy
- UTEX:
-
Culture Collection at the University of Texas at Austin, USA
References
Beijerinck, M. W. (1890). Culturversuche mit Zoochlorellen, Lichenengonidien und anderen niederen Algen. Botanische Zeitung, 47, 725–739, 741–754, 757–768, 781–785.
Bock, C., Krienitz, L., & Pröschold, T. (2011). Taxonomic reassessment of the genus Chlorella (Trebouxiophyceae) using molecular signatures (barcodes), including description of seven new species. Fottea, 11(2), 293–312. https://doi.org/10.5507/FOT.2011.028
Bock, C., Proschold, T., & Krienitz, L. (2010). Two new Dictyosphaerium-morphotype lineages of the Chlorellaceae (Trebouxiophyceae): Heynigia gen. nov. and Hindakia gen. nov. European Journal of Phycology, 45(3), 267–277. https://doi.org/10.1080/09670262.2010.487920
Bouckaert, R., Vaughan, T. G., Barido-Sottani, J., et al. (2019). BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLOS Computational Biology, 15(4), e1006650. https://doi.org/10.1371/journal.pcbi.1006650
Bradley, I. M., Pinto, A. J., & Guest, J. S. (2016). Design and evaluation of Illumina MiSeq compatible primers for the 18S rRNA gene for improved characterization of mixed microalgal communities. AEM, 82(19), 5878–5891. https://doi.org/10.1128/AEM.01630-16
Caisová, L., Marin, B., & Melkonian, M. (2013). A consensus secondary structure of ITS2 in the Chlorophyta identified by phylogenetic reconstruction. Protist, 164, 482–496. https://doi.org/10.1016/j.protis.2013.04.005
Chae, H., Lim, S., Kim, H., Choi, H.-G., & Kim, J. H. (2019). Morphology and phylogenetic relationships of Micractinium (Chlorellaceae, Trebouxiophyceae) taxa, including three new species from Antarctica. Algae, 34(4), 267–275. https://doi.org/10.4490/algae.2019.34.10.15
Coleman, A. W. (2000). The significance of a coincidence between evolutionary landmarks found in mating affinity and a DNA sequence. Protist, 151(1), 1–9. https://doi.org/10.1078/1434-4610-00002
Coleman, A. W. (2007). Pan-eukaryote ITS2 homologies revealed by RNA secondary structure. NAR, 35, 3322–3329. https://doi.org/10.1093/nar/gkm233
Coleman, A. W. (2009). Is there a molecular key to the level of ‘biological species’ in eukaryotes? A DNA guide. Molecular Phylogenetics and Evolution, 50, 197–203. https://doi.org/10.1016/j.ympev.2008.10.008
Coleman, A. W. (2015). Nuclear rRNA transcript processing versus internal transcribed spacer secondary structure. Trends in Genetics, 31(3), 157–163. https://doi.org/10.1016/j.tig.2015.01.002
Decelle, J., Romac, S., Sasaki, E., Not, F., Mahé, F., & Lovejoy, C. (2014). Intracellular diversity of the V4 and V9 regions of the 18S rRNA in marine protists (Radiolarians) assessed by high-throughput sequencing. PLoS ONE, 9(8), e104297. https://doi.org/10.1371/journal.pone.0104297
Drummond, A. J., & Rambaut, A. (2007). BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology, 7(1), 214. https://doi.org/10.1186/1471-2148-7-214
Egorova, I. N., Mincheva, E. V., & Boldina, O. N. (2018). Ataktogamous green microalgae of the genus Chlorosarcinopsis Herndon (Chlorophyceae, Chlorophyta) from Zabaikalskiy region (Russia). Phytotaxa, 343(1), 1–19. https://doi.org/10.11646/phytotaxa.343.1.1
Esteban, G. F., Fenchel, T., & Finlay, B. J. (2010). Mixotrophy in ciliates. Protist, 161, 621–641. https://doi.org/10.1016/j.protis.2010.08.002
Fawley, M. W., Fawley, K. P., & Owen, H. A. (2005). Diversity and ecology of small coccoid green algae from Lake Itasca, Minnesota, USA, including Meyerella planktonica, gen. et sp. nov. Phycologia, 44, 35–48. https://doi.org/10.2216/0031-8884(2005)44[35:DAEOSC]2.0.CO;2
Foissner, W. (2019). A detailed description of a Brazilian Holophrya teres (Ehrenberg, 1834) and nomenclatural revision of the genus Holophrya (Ciliophora, Prostomatida). European Journal of Protistology. https://doi.org/10.1016/j.ejop.2019.125662
Frolova, L. L., & Sverdrup, A. E. (2019). Diversity of Verhniy Kaban lake by 18S rRNA of hydrobionts on next-generation sequecing method. Bioscience Biotechnology Research Communications, 12(5), 329–335.
Fučíková, K., Lewis, P. O., & Lewis, L. A. (2014). Widespread desert affiliation of trebouxiophycean algae (Trebouxiophyceae, Chlorophyta) including discovery of three new desert genera. Phycological Research, 62(4), 294–305. https://doi.org/10.1111/pre.12062
Fujisawa, T., & Barraclough, T. G. (2013). Delimiting species using single-locus data and the generalized mixed Yule coalescent approach: A revised method and evaluation on simulated data sets. Systematic Biology, 62(5), 707–724. https://doi.org/10.1093/sysbio/syt033
Gaonkar, C. C., Piredda, R., Minucci, C., et al. (2018). Annotated 18S and 28S rDNA reference sequences of taxa in the planktonic diatom family Chaetocerotaceae. PLoS ONE, 13(12), e0208929. https://doi.org/10.1371/journal.pone.0208929
Garrido-Benavent, I., Pérez-Ortega, S., & de Los Ríos, A. (2017). From Alaska to Antarctica: Species boundaries and genetic diversity of Prasiola (Trebouxiophyceae), a foliose chlorophyte associated with the bipolar lichen-forming fungus Mastodia tessellate. Molecular Phylogenetics and Evolution, 107, 117–131. https://doi.org/10.1371/10.1016/u.ympev.2016.10.013
Guiry, M. D., & Guiry, G. M. (2021). AlgaeBase. World-wide electronic publication, National University of Ireland, Galway. Retrieved December 8, 2021, from http://www.algaebase.org
Heeg, J. S., & Wolf, M. (2015). ITS2 and 18S rDNA sequence-structure phylogeny of Chlorella and allies (Chlorophyta, Trebouxiophyceae, Chlorellaceae). Plant Gene, 4, 20–28. https://doi.org/10.1016/j.plgene.2015.08.001
Henley, W. J., Hironaka, J. L., Guillou, L., Buchheim, M. A., Buchheim, J. A., Fawley, M. W., & Fawley, K. P. (2004). Picochlorum oklahomensis gen. et sp. nov. (Trebouxiophyceae, Chlorophyta). Phycologia, 43, 641–652. https://doi.org/10.2216/i0031-8884-43-6-641.1
Hines, H. N., McCarthy, P. J., & Esteban, G. F. (2022). A Case Building Ciliate in the Genus Pseudoblepharisma Found in Subtropical Fresh Water. Diversity, 14, 174. https://doi.org/10.3390/d14030174
Hoshina R., & Fujiwara,Y. (2013). Molecular characterization of Chlorella cultures of the National Institute for Environment Studies culture collection with description of Micractinium inermum sp. nov., Didymogenes sphaerica sp. nov. and Didymogenes soliella sp. nov. (Chlorellaceae, Trebouxiophyceae). Phycological Research, 61(2), 124–132. https://doi.org/10.1111/pre.12010
Hoshina, R., Iwataki, M., & Imamura, N. (2010). Chlorella variabilis and Micractinium reisseri sp. nov. (Chlorellaceae, Trebouxiophyceae): Redescription of the endosymbiotic green algae of Paramecium bursaria (Peniculia, Oligohymenophorea) in the 120th year. Phycological Research, 58(3), 188–210. https://doi.org/10.1111/j.1440-1835.2010.00579.x
Hoshina, R., Kobayashi, M., Suzaki, T., & Kusuoka, Y. (2017). Brandtia ciliaticola gen. et sp. nov. (Chorellaceae, Trebouxiophyceae) a commom symbiotic green coccoid of various ciliate species. Phycological Research, 66(1), 76–81. https://doi.org/10.1111/pre.12194
Hoshina, R., & Nakada, T. (2018). Carolibrandtia nom. nov. as a replacement name for Brandtia Hoshina (Chlorellaceae, Trebouxiophyceae). Phycological Research, 66(1), 82–83. https://doi.org/10.1111/pre.12208
Hoshina, R., Tsukii, Y., Harumoto, T., & Suzaki, T. (2021). Characterization of a green Stentor with symbiotic algae growing in an extremely oligotrophic environment and storing large amounts of starch granules in its cytoplasm. Scientific Reports, 11, 2865. https://doi.org/10.1038/s41598-021-82416-9
Johnson, J. L., Fawley, M. W., & Fawley, K. P. (2007). The diversity of Scenedesmus and Desmodesmus (Chlorophyceae) in Itasa State Park, Minnesota, USA. Phycologia, 46, 214–229. https://doi.org/10.2216/05-69.1
Karpagam, R., Preeti, R., Jawahar, R.K., Saranya, S., Ashokkumar, B., & Varalakshmi, P. (2015). Fatty acid biosynthesis from a new isolate Meyerella sp. N4: molecular characterization, nutrient starvation, and fatty acid profiling for lipid enhancement. Energy & Fuels, 29(1), 143–149. https://doi.org/10.1021/ef501969a
Katana, A., Kwiatowski, J., Spalik, K., Zakryś, B., Szalacha, E., & Szymańska, H. (2001). Phylogenetic position of Koliella (Chlorophyta) as inferred from nuclear and chloroplast small subunit rDNA. Journal of Phycology, 37(3), 443–451. https://doi.org/10.1046/j.1529-8817.2001.037003443.x
Krasovec, M., Vancaester, E., Rombauts, S., et al. (2018). Genome analyses of the microalga Pichochlorum provide insights into the evolution of thermotolerance in the green lieage. Genome Biology and Evolution, 10(9), 2347–2365. https://doi.org/10.1093/gbe/evy167
Krienitz, L., Bock, C., Dadheech, P. K., & Proeschold, T. (2011). Taxonomic reassessment of the genus Mychonastes (Chlorophyceae, Chlorophyta) including the description of eight new species. Phycologia, 50(1), 89–106. https://doi.org/10.2216/10-15.1
Krienitz, L., Hegewald, E. H., Hepperle, D., Huss, V. A. R., Rohr, T., & Wolf, M. (2004). Phylogenetic relationship of Chlorella and Parachlorella gen. nov. (Chlorophyta, Trebouxiophyceae). Phycologia, 43, 529–542. https://doi.org/10.2216/i0031-8884-43-5-529.1
Krivina, E., & Temraleeva, A. (2020). The difficulty identifying and the cryptic diversity of Chlorella-clada microalgae (Chlorophyta). Microbiology, 89(6), 714–727. https://doi.org/10.1134/S0026261720060107
Krivina, E., Temraleeva, A., & Bukin, Y. S. (2021a). Species delimitation and cryptic diversity analysis of Parachlorella-clade microalgae (Chlorophyta). Microbiology, 90, 455–469. https://doi.org/10.1134/S0026261721040081
Krivina, E., Temraleeva, A., & Sinetova, A. (2021b). New species Micractinium kostikovii (Chlorellaceae, Trebouxiophyceae) from Russia. Phycological Research. https://doi.org/10.1111/pre.12469
Lambert, A., & Steel, M. (2013). Predicting the loss of phylogenetic diversity under non-stationary diversification models. Journal of Theoretical Biology, 337, 111–124. https://doi.org/10.1016/j.jtbi.2013.08.009
Lanzoni, O., Fokin, S. I., Lebedeva, N., Migunova, A., Petroni, G., & Potekhin, A. (2016). Rare freshwater ciliate Paramecium chlorelligerum Kahl, 1935 and its macronuclear symbiotic bacterium candidatus Holospora parva. PLoS ONE, 11(12), e0167928. https://doi.org/10.1371/journal.pone.0167928
Luo, W., Pflugmacher, S., Pröschold, T., Walz, N., & Krienitz, L. (2006). Genotype versus phenotype variability in Chlorella and Micractinium (Chlorophyta, Trebouxiophyceae). Protist, 157, 315–333. https://doi.org/10.1016/j.protis.2006.05.006
Malavasi, V., Škaloud, P., Rindi, F., Tempesta, S., Paoletti, M., Pasqualetti, M., & Fontaneto, D. (2016), DNA-based taxonomy in ecologically versatile microalgae: A re-evaluation of the species concept within the coccoid green algal genus Coccomyxa (Trebouxiophyceae, Chlorophyta). PLoS ONE, 11(3), e0151137. https://doi.org/10.1371/journal.pone.0151137
Nomokonova, V. I., Vykhristyuk, L. A., & Tarasova, N. G. (2001). Trophic state of Vasilievskiy lakes of Togliatti suburb. Izvestiya of the Samara Russian Academy of Sciences Scientific Center, 3(2), 274–283.
Paradis, E., Claude, J., & Strimmer, K. (2004). APE: Analyses of phylogenetics and evolution in R language. Bioinformatics, 20(2), 289–290. https://doi.org/10.1093/bioinformatics/btg412
Pröschold, T., Bock, C., Luo, W., & Krienitz, L. (2010). Polyphyletic distribution of bristle formation in Chlorellaceae: Micractinium, Diacanthos, Didymogenes and Hegewaldia gen. nov. (Trebouxiophyceae, Chlorophyta). Phycological Research, 58, 1–8. https://doi.org/10.1111/j.1440-1835.2009.00552.x
Pröschold, T., & Darienko, T. (2020). Choricystis and Lewiniosphaera gen. nov. (Trebouxiophyceae, Chlorophyta), two different green algal endosymbionts in freshwater sponges. Symbiosis, 82(3),1–14. https://doi.org/10.1007/s13199-020-00711-x
Pröschold, T., Darienko, T., Silva, P. C., Reisser, W., & Krienitz, L. (2011). The systematics of Zoochlorella revisited employing an integrative approach. Environmental Microbiology, 13, 350–364. https://doi.org/10.1111/j.1462-2920.2010.02333.x
Pröschold, T., Pitsch, G., & Darienko, T. (2020). Micractinium tetrahymenae (Trebouxiophyceae, Chlorophyta), a new endosymbiont isolated from Ciliates. Diversity, 12, 200. https://doi.org/10.3390/d12050200
Pröschold, T., Rieser, D., Darienko, T., et al. (2021). An integrative approach sheds new light onto the systematics and ecology of the widespread ciliate genus Coleps (Ciliophora, Prostomatea). Scientific Reports, 11, 5916. https://doi.org/10.1038/s41598-021-84265-y
Puillandre, N., Lambert, A., Brouillet, S., & Achaz, G. (2012). ABGD, automatic barcode gap discovery for primary species delimitation. Molecular Ecology, 21, 1864–1877. https://doi.org/10.1111/j.1365-294X.2011.05239.x
Seibel, P.N., Müller, T., Dandekar, T., Schultz, J., & Wolf, M. (2006). 4SALE: a tool for synchronous RNA sequence and secondary structure alignment and editing. BMC Bioinformatics, 7, 1−498. https://doi.org/10.1186/1471-2105-7-498
Seibel, P.N., Müller, T., Dandekar, T., & Wolf, M. (2008). Synchronous visual analysis and editing of RNA sequence and secondary structure alignments using 4SALE. BMC Research Notes, 1, 1−91. https://doi.org/10.1186/1756-0500-1-91
Simpson, P. D., & Van Valkenburg, S. D. (1978). The ultrastructure of Mychonastes ruminatus gen. et sp. nov., a new member of the Chlorophyceae isolated from brackish water. British Phycological Journal, 13(2), 117–130. https://doi.org/10.2216/10-15.1
Sogin, M. L., Morrison, H. G., Huber, J. A., et al. (2006). Microbial diversity in the deep sea and the underexplored rare biosphere. Proceedings of the National Academy of Sciences, 103, 12115–12120. https://doi.org/10.1073/pnas.0605127103
Somogyi, B., Felföldi, T., Solymosi, K., Flieger, K., Márialigeti, K., Böddi, B., & Vörös, L. (2014). One step closer to eliminating the nomenclatural problems of minute coccoid green algae: Pseudochloris wilhelmii, gen. et sp. nov. (Trebouxiophyceae, Chlorophyta). European Journal of Phycology, 48(4), 427–436. https://doi.org/10.1080/09670262.2013.854411
Spanner, C., Darienko, T., Biehler, T., Sonntag, B., & Pröschold, T. (2020). Endosymbiotic green algae in Paramecium bursaria: A new isolation method and a simple diagnostic PCR approach for the identification. Diversity, 12(6), 240. https://doi.org/10.3390/d12060240
Stanier, R. Y., Kunisawa, R., Mandel, M., & Cohen-Bazire, G. (1971). Purification and properties of unicellular blue-green algae (order Chroococcales). Bacteriology Reviews, 35, 171–205. https://doi.org/10.1128/br.35.2.171-205.1971
Sverdrup, A. E., & Frolova, L. L. (2020). Saprobity identification of hydrobiont species of Verhniy Kaban lake of Kazan by 18S rRNA marker gene. Scientific Notes of V.I. Vernadsky Crimean Federal University. Biology Chemistry, 4, 127–142. https://doi.org/10.37279/2413-1725-2020-6-4-127-142
Temraleeva, A., Krivina, E., & Boldina, O. (2022) Edaphochloris gen. nov.: A new genus of soil green algae (Trebouxiophyceae, Chlorophyta) with simple morphology. Plant Systematics and Evolution. https://doi.org/10.1007/s00606-021-01795-8
Temraleeva, A., Moskalenko, S., Mincheva, E., Bukin, Y., & Sinetova, M. (2018). Spongiosarcinopsis terrestris gen. et sp. nov. (Chlorophyta, Chlorophyceae): A new genus of green algae from gray forest soil, Russia. Phytotaxa, 376(6), 291–300. https://doi.org/10.11646/phytotaxa.376.6.4
Vorobyev, K., Andronov, E., Rautian, M., Skoblo, I., Migunova, A., & Kvitko, K. (2009). An atypical Chlorella symbiont from Paramecium bursaria. Protistology, 6(1), 39–44.
White, T. J., Bruns, T., Lee, S., & Taylor, J. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, & T. J. White (Eds.), PCR Protocols: A Guide to Methods and Applications (pp. 315–322). Academic Press.
Zou, S., Fei, C., Song, J., Bao, Y., He, M., & Wang, C. (2016a). Combining and comparing coalescent, distance and character-based approaches for barcoding microalgaes: A test with Chlorella-like species (Chlorophyta). PloS O, 11(4), e0153833. https://doi.org/10.1371/journal.pone.0153833
Zou, S., Fei, C., Wang, C., Gao, Z., Bao, Y., He, M., & Wang, C. (2016b). How DNA barcoding can be more effective in microalgae identification: A case of cryptic diversity revelation in Scenedesmus (Chlorophyceae). Science and Reports, 9(6), 36822. https://doi.org/10.1038/srep36822
Acknowledgements
We thank the Bioline company for the opportunity to use of the Leica THUNDER 3D Cell Culture Imaging system (Leica Microsystems, Germany).
Funding
The work was funded by the Russian Foundation for Basic Research (RFBR), project no.19–34-60002 (sampling, isolation, cultivation and morphological observation, DNA extraction, PCR, phylogenetic analysis) and the Ministry of Science and Higher Education of the Russian Federation project no.0279–2021-0010 (species delimitation), project no.N121021600184-6 (ultrastructural observations by transmission electron microscopy), project no.121040800126–5 (maintaining algae strains in the ACSSI collection).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Human and animals participation
Our research did not involve human or animals participants.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Krivina, E.S., Boldina, O.N., Bukin, Y.S. et al. Species delimitation polyphasic approach reveals Meyerella similis sp. nov.: a new species of “small green balls” within the Chlorella-clade (Trebouxiophyceae, Chlorophyta). Org Divers Evol 23, 25–40 (2023). https://doi.org/10.1007/s13127-022-00590-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13127-022-00590-8