Abstract
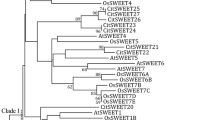
Hydroxycinnamate-CoA quinate hydroxycinnamoyl transferase (HQT) enzyme affect plant secondary metabolism and are crucial for growth and development. To date, limited research on the genome-wide analysis of HQT family genes and their regulatory roles in chlorogenic acid (CGA) accumulation in leafy vegetable sweet potato is available. Here, a total of 58 HQT family genes in the sweet potato genome (named IbHQT) were identified and analyzed. We studied the chromosomal distribution, phylogenetic relationship, motifs distribution, collinearity, and cis-acting element analysis of HQT family genes. This study used two sweet potato varieties, high CGA content Fushu 7-6-14-7 (HC), and low CGA content Fushu 7-6 (LC). Based on the phylogenetic analysis, clade A was unique among the identified four clades as it contained HQT genes from various species. The chromosomal location and collinearity analysis revealed that tandem gene duplication may promote the IbHQT gene expansion and expression. The expression patterns and profile analysis showed changes in gene expression levels at different developmental stages and under cold, drought, and salt stress conditions. The expression analysis verified by qRT-PCR revealed that IbHQT genes were highly expressed in the HC variety leaves than in the LC variety. Furthermore, cloning and gene function analysis unveiled that IbHQT family genes are involved in the biosynthesis and accumulation of CGA in sweet-potato. This study expands our understanding of the regulatory role of HQT genes in sweet-potato and lays a foundation for further functional characterization and genetic breeding by engineering targeted HQT candidate genes in various sweet-potato varieties and other species.








Similar content being viewed by others
References
Arnold PA, Kruuk LEB, Nicotra AB (2019) How to analyse plant phenotypic plasticity in response to a changing climate. New Phytol 222:1235–1241. https://doi.org/10.1111/nph.15656
Atayee AR, Noori MS (2020) Alleviation of cold stress in vegetable crops. J Sci Agric 4:38–44. https://doi.org/10.25081/jsa.2020.v4.6110
Cardenas CL, Costa MA, Laskar DD et al (2021) RNA i modulation of chlorogenic acid and lignin deposition in nicotiana tabacum and insufficient compensatory metabolic cross-talk. J Nat Prod 84:694–706. https://doi.org/10.1021/acs.jnatprod.1c00054
Chang J, Luo J, He G (2009) Regulation of polyphenols accumulation by combined overexpression/silencing key enzymes of phyenylpropanoid pathway. Acta Biochim Biophys Sin (shanghai) 41:123–130. https://doi.org/10.1093/abbs/gmn014
Cheevarungnapakul K, Khaksar G, Panpetch P et al (2019) Identification and functional characterization of genes involved in the biosynthesis of caffeoylquinic acids in sunflower (Helianthus annuus L.). Front Plant Sci. https://doi.org/10.3389/fpls.2019.00968
Chen Z, Tang N, You Y et al (2015) Transcriptome analysis reveals the mechanism underlying the production of a high quantity of chlorogenic acid in young leaves of lonicera macranthoides Hand.-Mazz. PLoS ONE 10:1–18. https://doi.org/10.1371/journal.pone.0137212
Cheng AX, Gou JY, Yu XH et al (2013) Characterization and ectopic expression of a populus hydroxyacid hydroxycinnamoyltransferase. Mol Plant 6:1889–1903. https://doi.org/10.1093/mp/sst085
Claude SJ, Park S, Park SJ (2021) Transcriptome-wide identification and quantification of caffeoylquinic acid biosynthesis pathway and prediction of its putative bahds gene complex in a. Spathulifolius. Int J Mol Sci. https://doi.org/10.3390/ijms22126333
Clé C, Hill LM, Niggeweg R et al (2008) Modulation of chlorogenic acid biosynthesis in Solanum lycopersicum; consequences for phenolic accumulation and UV-tolerance. Phytochemistry 69:2149–2156. https://doi.org/10.1016/j.phytochem.2008.04.024
Comino C, Hehn A, Moglia A et al (2009) The isolation and mapping of a novel hydroxycinnamoyltransferase in the globe artichoke chlorogenic acid pathway. BMC Plant Biol 9:1–13. https://doi.org/10.1186/1471-2229-9-30
D’Auria JC (2006) Acyltransferases in plants: a good time to be BAHD. Curr Opin Plant Biol 9:331–340. https://doi.org/10.1016/j.pbi.2006.03.016
Djami-Tchatchou AT, Matsaunyane LBT, Kalu CM, Ntushelo K (2019) Gene expression and evidence of coregulation of the production of some metabolites of chilli pepper inoculated with Pectobacterium carotovorum ssp. carotovorum. Funct Plant Biol 46:1114–1122. https://doi.org/10.1071/FP18244
Fazal H, Abbasi BH, Ahmad N et al (2016) Correlation of different spectral lights with biomass accumulation and production of antioxidant secondary metabolites in callus cultures of medicinally important Prunella vulgaris L. J Photochem Photobiol B Biol 159:1–7. https://doi.org/10.1016/j.jphotobiol.2016.03.008
Huang J, Sun W, Ren J et al (2020) Genome-wide identification and characterization of actin-depolymerizing factor (Adf) family genes and expression analysis of responses to various stresses in zea mays l. Int J Mol Sci 21:1–17. https://doi.org/10.3390/ijms21051751
Jo Y, Kim SM, Choi H et al (2020) Sweet potato viromes in eight different geographical regions in Korea and two different cultivars. Sci Rep 10:1–16. https://doi.org/10.1038/s41598-020-59518-x
Jung JK, Lee SU, Kozukue N et al (2011) Distribution of phenolic compounds and antioxidative activities in parts of sweet potato (Ipomoea batata L.) plants and in home processed roots. J Food Compos Anal 24:29–37. https://doi.org/10.1016/j.jfca.2010.03.025
Klopfenstein DV, Zhang L, Pedersen BS et al (2018) GOATOOLS: a python library for gene ontology analyses. Sci Rep 8:1–17. https://doi.org/10.1038/s41598-018-28948-z
Lallemand LA, Zubieta C, Lee SG et al (2012) A structural basis for the biosynthesis of the major chlorogenic acids found in coffee. Plant Physiol 160:249–260. https://doi.org/10.1104/pp.112.202051
Laurie SM, Bairu MW (2022) Analysis of the nutritional composition and drought tolerance traits of sweet potato : selection criteria for breeding lines. Plants. https://doi.org/10.3390/plants11141804
Lepelley M, Cheminade G, Tremillon N et al (2007) Chlorogenic acid synthesis in coffee: An analysis of CGA content and real-time RT-PCR expression of HCT, HQT, C3H1, and CCoAOMT1 genes during grain development in C. canephora. Plant Sci 172:978–996. https://doi.org/10.1016/j.plantsci.2007.02.004
Li G, Yu S, Yang XS, Chen QF (2011) Study on extraction technology for chlorogenic acid from sweet potato leaves by orthogonal design. Procedia Environ Sci 8:403–407. https://doi.org/10.1016/j.proenv.2011.10.063
Li J, Wang T, Han J, Ren Z (2020) Genome-wide identification and characterization of cucumber bHLH family genes and the functional characterization of CsbHLH041 in NaCl and ABA tolerance in Arabidopsis and cucumber. BMC Plant Biol 20:1–20. https://doi.org/10.1186/s12870-020-02440-1
Li K, Duan L, Zhang Y et al (2021) Genome-wide identification and expression profile analysis of trihelix transcription factor family genes in response to abiotic stress in sorghum [Sorghum bicolor (L.) Moench]. BMC Genomics 22:1–17. https://doi.org/10.1186/s12864-021-08000-7
Liu X, Zhang H, Ma L et al (2020) Genome-wide identification and expression profiling analysis of the trihelix gene family under abiotic stresses in medicago truncatula. Genes (Basel) 11:1–18. https://doi.org/10.3390/genes11111389
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:1–21. https://doi.org/10.1186/s13059-014-0550-8
Luo D, Mu T, Sun H (2021) Profiling of phenolic acids and flavonoids in sweet potato (Ipomoea batatas L.) leaves and evaluation of their anti-oxidant and hypoglycemic activities. Food Biosci 39:100801. https://doi.org/10.1016/j.fbio.2020.100801
Moglia A, Acquadro A, Eljounaidi K et al (2016) Genome-wide identification of bahd acyltransferases and in vivo characterization of HQT-like enzymes involved in caffeoylquinic acid synthesis in globe artichoke. Front Plant Sci 7:1–15. https://doi.org/10.3389/fpls.2016.01424
Mudau SP, Steenkamp PA, Piater LA et al (2018) Metabolomics-guided investigations of unintended effects of the expression of the hydroxycinnamoyl quinate hydroxycinnamoyltransferase (hqt1) gene from Cynara cardunculus var. scolymus in Nicotiana tabacum cell cultures. Plant Physiol Biochem 127:287–298. https://doi.org/10.1016/j.plaphy.2018.04.005
Naveed M, Hejazi V, Abbas M et al (2018) Chlorogenic acid (CGA): a pharmacological review and call for further research. Biomed Pharmacother 97:67–74. https://doi.org/10.1016/j.biopha.2017.10.064
Niggeweg R, Michael AJ, Martin C (2004) Engineering plants with increased levels of the antioxidant chlorogenic acid. Nat Biotechnol 22:746–754. https://doi.org/10.1038/nbt966
Pan C, Wang S, Yang C et al (2022a) Genome-wide identification and expression profiling analysis of Wnt family genes affecting adipocyte differentiation in cattle. Sci Rep 12:1–14. https://doi.org/10.1038/s41598-021-04468-1
Pan R, Buitrago S, Peng Y et al (2022b) Genome-wide identification of cold-tolerance genes and functional analysis of IbbHLH116 gene in sweet potato. Gene 837:146690. https://doi.org/10.1016/j.gene.2022.146690
Park SU, Kim YH, Lee CJ et al (2020) Comparative transcriptome profiling of two sweetpotato cultivars with contrasting flooding stress tolerance levels. Plant Biotechnol Rep 14:743–756. https://doi.org/10.1007/s11816-020-00650-5
Payyavula RS, Shakya R, Sengoda VG et al (2015) Synthesis and regulation of chlorogenic acid in potato: Rerouting phenylpropanoid flux in HQT-silenced lines. Plant Biotechnol J 13:551–564. https://doi.org/10.1111/pbi.12280
Peng X, Li W, Wang W, Bai G (2010) Cloning and characterization of a cDNA coding a hydroxycinnamoyl-CoA quinate hydroxycinnamoyl transferase involved in chlorogenic acid biosynthesis in lonicera japonica. Planta Med 76:1921–1926. https://doi.org/10.1055/s-0030-1250020
Qin N, Gao Y, Cheng X et al (2021) Genome-wide identification of CLE gene family and their potential roles in bolting and fruit bearing in cucumber (Cucumis sativus L.). BMC Plant Biol 21:1–18. https://doi.org/10.1186/s12870-021-02900-2
Siahpoush A, Ghasemi N, Shams Ardakani M, Asghari G (2011) Production of chlorogenic acid in Varthemia persica DC (var. persica) callus cultures. Res Pharm Sci 6:63–69
Sonnante G, D’Amore R, Blanco E et al (2010) Novel hydroxycinnamoyl-coenzyme a quinate transferase genes from artichoke are involved in the synthesis of chlorogenic acid. Plant Physiol 153:1224–1238. https://doi.org/10.1104/pp.109.150144
Tuominen LK, Johnson VE, Tsai CJ (2011) Differential phylogenetic expansions in BAHD acyltransferases across five angiosperm taxa and evidence of divergent expression among Populus paralogues. BMC Genomics. https://doi.org/10.1186/1471-2164-12-236
Ullah MA, Tungmunnithum D, Garros L et al (2019) Monochromatic lights-induced trends in antioxidant and antidiabetic polyphenol accumulation in in vitro callus cultures of Lepidium sativum L. J Photochem Photobiol B Biol 196:111505. https://doi.org/10.1016/j.jphotobiol.2019.05.002
Wang F, Tong W, Zhu H et al (2016) A novel Cys2/His2 zinc finger protein gene from sweetpotato, IbZFP1, is involved in salt and drought tolerance in transgenic Arabidopsis. Planta 243:783–797. https://doi.org/10.1007/s00425-015-2443-9
Wei Q, Lan K, Liu Y et al (2022) Transcriptome analysis reveals regulation mechanism of methyl jasmonate-induced terpenes biosynthesis in Curcuma wenyujin. PLoS ONE 17:e0270309. https://doi.org/10.1371/journal.pone.0270309
Xu Y, Tie W, Yan Y et al (2021) Identification and expression of the BAHD family during development, ripening, and stress response in banana. Mol Biol Rep 48:1127–1138. https://doi.org/10.1007/s11033-020-06132-9
Yan K, Cui M, Zhao S et al (2016) Salinity stress is beneficial to the accumulation of chlorogenic acids in honeysuckle (Lonicera Japonica Thunb.). Front Plant Sci 7:1–10. https://doi.org/10.3389/fpls.2016.01563
Yang Z, Sun J, Chen Y et al (2019) Genome-wide identification, structural and gene expression analysis of the bZIP transcription factor family in sweet potato wild relative Ipomoea trifida. BMC Genet 20:1–18. https://doi.org/10.1186/s12863-019-0743-y
Zhang J, Lin LM, Cheng WW et al (2021) Genome-wide identification and expression analysis of glycosyltransferase gene family 1 in Quercus robur L. J Appl Genet 62:559–570. https://doi.org/10.1007/s13353-021-00650-3
Zhang J, Wu M, Li W, Bai G (2017) Regulation of chlorogenic acid biosynthesis by hydroxycinnamoyl CoA quinate hydroxycinnamoyl transferase in Lonicera japonica. Plant Physiol Biochem 121:74–79. https://doi.org/10.1016/j.plaphy.2017.10.017
Zhao K, Chen S, Yao W et al (2021) Genome-wide analysis and expression profile of the bZIP gene family in poplar. BMC Plant Biol 21:1–16. https://doi.org/10.1186/s12870-021-02879-w
Funding
This work was supported by the National Key R & D Program of China (Grant Nos. 2018YFD1000700, 2018YFD1000705, 2019YFD1001300, 2019YFD1001305).
Author information
Authors and Affiliations
Contributions
MBM, investigation, validation, writing-original draft, writing-review & editing; RP, formal analysis, validation, writing-review & editing; YP, validation; RGM, validation, writing-original draft, writing-review & editing; AS, writing-review & editing; XSY, Resources, funding acquisition; WZ, conceptualization, writing-review & editing, funding acquisition, and supervision. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
12298_2023_1299_MOESM5_ESM.tif
Supplementary Fig. S1 Conserved motifs analysis of ‘DVJKKALAKALVSYYPLAGRL’ sequence denoted by serial number ‘motif 4’ (TIF 1950 KB)
12298_2023_1299_MOESM6_ESM.tif
Supplementary Fig. S2 Analysis of cis-acting elements in the promoter region of IbHQT genes. A large number of cis-acting elements are represented in different colors and the elements are shown on the right (TIF 5052 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Medison, M.B., Pan, R., Peng, Y. et al. Identification of HQT gene family and their potential function in CGA synthesis and abiotic stresses tolerance in vegetable sweet potato. Physiol Mol Biol Plants 29, 361–376 (2023). https://doi.org/10.1007/s12298-023-01299-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12298-023-01299-4