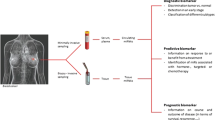
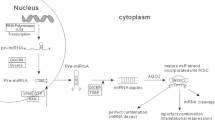
Abstract
MicroRNAs (miRNAs) negatively affect gene expression by binding to their specific mRNAs resulting in either mRNA destruction or translational repression. The aberrant expression of various miRNAs has been associated with a number of human cancer. Oncogenic or tumor-suppressor miRNAs regulate a variety of pathways involved in the development of breast cancer (BC), including cell proliferation, apoptosis, metastasis, cancer recurrence, and chemoresistance. Variations in miRNA-encoding genes and their target genes lead to dysregulated gene expression resulting in the development and progression of BC. The various therapeutic approaches to treat the disease include chemotherapy, radiation therapy, surgical removal, hormone therapy, chemotherapy, and targeted biological therapy. The purpose of the current review is to explore the genetic variations in tumor-suppressor miRNA-encoding genes and their target genes in association with the disease development and prognosis. The therapeutic interventions targeting the variants for better disease outcomes have also been discussed.


Similar content being viewed by others
Data availability
Not applicable.
References
Poorkiani M, Abbaszadeh A, Hazrati M, Jafari P, Sadeghi M, Mohammadianpanah M. The effect of rehabilitation on quality of life in female breast cancer survivors in Iran. Indian J Med Paediatric Oncol. 2010;31(4):105–9.
Kashyap D, Pal D, Sharma R, Garg VK, Goel N, Koundal D, et al. Global increase in breast cancer incidence: risk factors and preventive measures. Biomed Res Int. 2022;2022:9605439.
Konieczny M, Cipora E, Sygit K, Fal A. Quality of life of women with breast cancer and socio-demographic factors. Asian Pac J Cancer Prevention. 2020;21(1):185–93.
Lee EY, Muller WJ. Oncogenes and tumor suppressor genes. Cold Spring Harb Perspect Biol. 2010;2(10): a003236.
Choudhuri S, Cui Y, Klaassen CD. Molecular targets of epigenetic regulation and effectors of environmental influences. Toxicol Appl Pharmacol. 2010;245(3):378–93.
Zhang Y, Zhang L, Wang Y, Ding H, Xue S, Qi H, et al. MicroRNAs or long noncoding RNAs in diagnosis and prognosis of coronary artery disease. Aging Dis. 2019;10(2):353–66.
Chacon-Cortes D, Smith RA, Haupt LM, Lea RA, Youl PH, Griffiths LR. Genetic association analysis of miRNA SNPs implicates MIR145 in breast cancer susceptibility. BMC Med Genet. 2015;16:107–107.
Palmero EI, de Campos SG, Campos M, de Souza NC, Guerreiro ID, Carvalho AL, et al. Mechanisms and role of microRNA deregulation in cancer onset and progression. Genet Mol Biol. 2011;34(3):363–70.
Gu TJ, Yi X, Zhao XW, Zhao Y, Yin JQ. Alu-directed transcriptional regulation of some novel miRNAs. BMC Genom. 2009;10:563.
Loh HY, Norman BP, Lai KS, Rahman N, Alitheen NBM, Osman MA. The regulatory role of MicroRNAs in breast cancer. Int J Mol Sci. 2019;20(19):4940.
Dai ZM, Kang HF, Zhang WG, Li HB, Zhang SQ, Ma XB, et al. The associations of single nucleotide polymorphisms in miR196a2, miR-499, and miR-608 with breast cancer susceptibility: a STROBE-compliant observational study. Medicine (Baltimore). 2016;95(7): e2826.
Ye D, Shen Z, Zhou S. Function of microRNA-145 and mechanisms underlying its role in malignant tumor diagnosis and treatment. Cancer Manag Res. 2019;11:969–79.
Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci. 2004;101(9):2999–3004.
Shi B, Sepp-Lorenzino L, Prisco M, Linsley P, DeAngelis T, Baserga R. Micro RNA 145 targets the insulin receptor substrate-1 and inhibits the growth of colon cancer cells. J Biol Chem. 2007;282(45):32582–90.
Kontorovich T, Levy A, Korostishevsky M, Nir U, Friedman E. Single nucleotide polymorphisms in miRNA binding sites and miRNA genes as breast/ovarian cancer risk modifiers in Jewish high-risk women. Int J Cancer. 2010;127(3):589–97.
Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137(4):647–58.
Cho WC, Chow AS, Au JS. MiR-145 inhibits cell proliferation of human lung adenocarcinoma by targeting EGFR and NUDT1. RNA Biol. 2011;8(1):125–31.
Yan X, Chen X, Liang H, Deng T, Chen W, Zhang S, et al. miR-143 and miR-145 synergistically regulate ERBB3 to suppress cell proliferation and invasion in breast cancer. Mol Cancer. 2014;13(1):1–14.
Chacon-Cortes D, Smith RA, Lea RA, Youl PH, Griffiths LR. Association of microRNA 17–92 cluster host gene (MIR17HG) polymorphisms with breast cancer. Tumor Biol. 2015;36(7):5369–76.
Duan X, Zhang D, Wang S, Feng X, Wang T, Wang P, et al. Effects of polycyclic aromatic hydrocarbon exposure and miRNA variations on peripheral blood leukocyte DNA telomere length: a cross-sectional study in Henan Province, China. Sci Total Environ. 2020;703: 135600.
Wang D, Liu Q, Ren Y, Zhang Y, Wang X, Liu B. Association analysis of miRNA-related genetic polymorphisms in miR-143/145 and KRAS with colorectal cancer susceptibility and survival. Biosci Rep 2021;41(4):BSR20204136.
Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010;17(2):193–9.
Rokavec M, Li H, Jiang L, Hermeking H. The p53/miR-34 axis in development and disease. J Mol Cell Biol. 2014;6(3):214–30.
Bhat SA, Majid S, Hassan T. MicroRNAs and its emerging role as breast cancer diagnostic marker—a review. Adv Biomark Sci Technol. 2019;1:1–8.
Hahn S, Jackstadt R, Siemens H, Hünten S, Hermeking H. SNAIL and miR-34a feed-forward regulation of ZNF281/ZBP99 promotes epithelial-mesenchymal transition. Embo J. 2013;32(23):3079–95.
Imani S, Wei C, Cheng J, Khan MA, Fu S, Yang L, et al. MicroRNA-34a targets epithelial to mesenchymal transition-inducing transcription factors (EMT-TFs) and inhibits breast cancer cell migration and invasion. Oncotarget. 2017;8(13):21362–79.
Okada N, Lin CP, Ribeiro MC, Biton A, Lai G, He X, et al. A positive feedback between p53 and miR-34 miRNAs mediates tumor suppression. Genes Dev. 2014;28(5):438–50.
Venkatesan, N, Xavier A, Sindhu KJ, Sinha H, Devarajan K, Identification and validation of putative target genes regulated by <em>miR-34</em> in cervical cancer. bioRxiv, 2021: p. 2021.09.02.458804.
Tsiakou A, Zagouri F, Zografos E, Samelis G, Gazouli M, Kalapanida D, et al. Prognostic significance of miR-34 rs4938723 T> C polymorphism in triple negative breast cancer patients. Clin Biochem. 2019;68:9–14.
Sanaei S, Hashemi M, Rezaei M, Hashemi SM, Bahari G, Ghavami S. Evaluation of the pri-miR-34b/c rs4938723 polymorphism and its association with breast cancer risk. Biomed Rep. 2016;5(1):125–9.
Bensen JT, Tse CK, Nyante SJ, Barnholtz-Sloan JS, Cole SR, Millikan RC. Association of germline microRNA SNPs in pre-miRNA flanking region and breast cancer risk and survival: the Carolina Breast Cancer Study. Cancer Causes Control. 2013;24(6):1099–109.
Yang H, Xu Z, Peng Y, Wang J, Xiang Y. Integrin β4 as a potential diagnostic and therapeutic tumor marker. 2021;11(8):1197.
Brendle A, Lei H, Brandt A, Johansson R, Enquist K, Henriksson R, et al. Polymorphisms in predicted microRNA-binding sites in integrin genes and breast cancer: ITGB4 as prognostic marker. Carcinogenesis. 2008;29(7):1394–9.
Xiao Y, Humphries B, Yang C, Wang Z. MiR-205 dysregulations in breast cancer: the complexity and opportunities. Noncoding RNA 2019;5(4).
Wu H, Zhu S, Mo YY. Suppression of cell growth and invasion by miR-205 in breast cancer. Cell Res. 2009;19(4):439–48.
Piovan C, Palmieri D, Di Leva G, Braccioli L, Casalini P, Nuovo G, et al. Oncosuppressive role of p53-induced miR-205 in triple negative breast cancer. Mol Oncol. 2012;6(4):458–72.
Qu S, Wang T, Huang J. Presence of the minor allele of microRNA205 rs3842530 polymorphism increases 18FDG uptake in patients with breast cancer via targeting VEGF. Mol Med Rep. 2018;17(1):636–42.
Thammaiah CK, Jayaram S. Role of let-7 family microRNA in breast cancer. Noncoding RNA Res. 2016;1(1):77–82.
Wang X, Cao L, Wang Y, Wang X, Liu N, You Y. Regulation of let-7 and its target oncogenes (review). Oncol Lett. 2012;3(5):955–60.
Sun H, Ding C, Zhang H, Gao J. Let-7 miRNAs sensitize breast cancer stem cells to radiation-induced repression through inhibition of the cyclin D1/Akt1/Wnt1 signaling pathway. Mol Med Rep. 2016;14(4):3285–92.
Ning Y, Xu M, Cao X, Chen X, Luo X. Inactivation of AKT, ERK and NF-κB by genistein derivative, 7-difluoromethoxyl-5, 4’-di-n-octylygenistein, reduces ovarian carcinoma oncogenicity. Oncol Rep. 2017;38(2):949–58.
Sun R, Gong J, Li J, Ruan Z, Yang X, Zheng Y et al. A genetic variant rs13293512 in the promoter of let-7 is associated with an increased risk of breast cancer in Chinese women. Biosci Rep 2019;39(5):BSR20182079.
Jančík S, Drábek J, Radzioch D, Hajdúch M. Clinical relevance of KRAS in human cancers. J Biomed Biotechnol 2010;2010:150960.
Chetty R, Govender D. Gene of the month: KRAS. Pancreas. 2013;57:90.
Paranjape T, Heneghan H, Lindner R, Keane FK, Hoffman A, Hollestelle A, et al. A 3’-untranslated region KRAS variant and triple-negative breast cancer: a case-control and genetic analysis. Lancet Oncol. 2011;12(4):377–86.
Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139(4):693–706.
Sanaei S, Hashemi M, Eskandari E, Hashemi SM, Bahari G. KRAS gene polymorphisms and their impact on breast cancer risk in an Iranian population. Asian Pac J Cancer Prev. 2017;18(5):1301–5.
Nogueira A, Fernandes M, Catarino R, Medeiros R. RAD52 functions in homologous recombination and its importance on genomic integrity maintenance and cancer therapy. Cancers. 2019;11(11):1622.
Jiang Y, Qin Z, Hu Z, Guan X, Wang Y, He Y, et al. Genetic variation in a hsa-let-7 binding site in RAD52 is associated with breast cancer susceptibility. Carcinogenesis. 2013;34(3):689–93.
Cao J, Luo C, Peng R, Guo Q, Wang K, Wang P, et al. MiRNA-binding site functional polymorphisms in DNA repair genes RAD51, RAD52, and XRCC2 and breast cancer risk in Chinese population. Tumor Biol. 2016;37:16039–51.
Adams BD, Furneaux H, White BA. The micro-ribonucleic acid (miRNA) miR-206 targets the human estrogen receptor-alpha (ERalpha) and represses ERalpha messenger RNA and protein expression in breast cancer cell lines. Mol Endocrinol. 2007;21(5):1132–47.
Chen X, Yan Q, Li S, Zhou L, Yang H, Yang Y, et al. Expression of the tumor suppressor miR-206 is associated with cellular proliferative inhibition and impairs invasion in ERα-positive endometrioid adenocarcinoma. Cancer Lett. 2012;314(1):41–53.
Chhichholiya Y, Suryan AK, Suman P, Munshi A, Singh S. SNPs in miRNAs and target sequences: role in cancer and diabetes. Front Genet. 2021;12: 793523.
Brucker SY, Frank L, Eisenbeis S, Henes M, Wallwiener D, Riess O, et al. Sequence variants in ESR1 and OXTR are associated with Mayer-Rokitansky-Küster-Hauser syndrome. Acta Obstet Gynecol Scand. 2017;96(11):1338–46.
Shi X-B, Xue L, Yang J, Ma A-H, Zhao J, Xu M, et al. An androgen-regulated miRNA suppresses Bak1 expression and induces androgen-independent growth of prostate cancer cells. Proc Natl Acad Sci. 2007;104(50):19983–8.
Zhang Y, Gao J-S, Tang X, Tucker LD, Quesenberry P, Rigoutsos I, et al. MicroRNA 125a and its regulation of the p53 tumor suppressor gene. FEBS Lett. 2009;583(22):3725–30.
Guo X, Wu Y, Hartley R. MicroRNA-125a represses cell growth by targeting HuR in breast cancer. RNA Biol. 2009;6(5):575–83.
Gaur A, Jewell DA, Liang Y, Ridzon D, Moore JH, Chen C, et al. Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res. 2007;67(6):2456–68.
Liang Z, Pan Q, Zhang Z, Huang C, Yan Z, Zhang Y, et al. MicroRNA-125a-5p controls the proliferation, apoptosis, migration and PTEN/MEK1/2/ERK1/2 signaling pathway in MCF-7 breast cancer cells. Mol Med Rep. 2019;20(5):4507–14.
Wang Y, Zeng G, Jiang Y. The emerging roles of miR-125b in cancers. Cancer Manag Res. 2020;12:1079–88.
Lehmann TP, Korski K, Ibbs M, Zawierucha P, Grodecka-Gazdecka S, Jagodziński PP. rs12976445 variant in the pri-miR-125a correlates with a lower level of hsa-miR-125a and ERBB2 overexpression in breast cancer patients. Oncol Lett. 2013;5(2):569–73.
Scott GK, Goga A, Bhaumik D, Berger CE, Sullivan CS, Benz CC. Coordinate suppression of ERBB2 and ERBB3 by enforced expression of micro-RNA miR-125a or miR-125b. J Biol Chem. 2007;282(2):1479–86.
Schulman BR, Esquela-Kerscher A, Slack FJ. Reciprocal expression of lin-41 and the microRNAs let-7 and mir-125 during mouse embryogenesis. Dev Dyn. 2005;234(4):1046–54.
Jo H, Shim K, Jeoung D. Potential of the miR-200 family as a target for developing anti-cancer therapeutics. Int J Mol Sci. 2022;23(11):5881.
Valastyan S, Reinhardt F, Benaich N, Calogrias D, Szász AM, Wang ZC, et al. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell. 2009;137(6):1032–46.
Valastyan S, Weinberg RA. miR-31: a crucial overseer of tumor metastasis and other emerging roles. Cell Cycle. 2010;9(11):2124–9.
Schunkert EM, Zhao W, Zänker K. Breast cancer recurrence risk assessment: is non-invasive monitoring an option? Biomed Hub. 2018;3(3):1–17.
Miller KD, Nogueira L, Mariotto AB, Rowland JH, Yabroff KR, Alfano CM, et al. 2019 Cancer treatment and survivorship statistics. CA. 2019;69(5):363–85.
Loh HY, Norman BP, Lai KS, Rahman N. The regulatory role of MicroRNAs in breast cancer. Int J Mol Sci. 2019;20(19):4940.
Banerjee B, Sherwood RI. A CRISPR view of gene regulation. Curr Opin Syst Biol. 2017;1:1–8.
Karn V, Sandhya S, Hsu W, Parashar D, Singh HN, Jha NK, et al. CRISPR/Cas9 system in breast cancer therapy: advancement, limitations and future scope. Cancer Cell Int. 2022;22(1):234.
Cong L, Zhang F. Genome engineering using CRISPR-Cas9 system. Methods Mol Biol. 2015;1239:197–217.
Tian Z, Liang G, Cui K, Liang Y, Wang Q, Lv S, et al. Insight into the prospects for RNAi therapy of cancer. Front Pharmacol 2021;12:644718.
Bottai G, Truffi M, Corsi F, Santarpia L. Progress in nonviral gene therapy for breast cancer and what comes next? Expert Opin Biol Ther. 2017;17(5):595–611.
Ngamcherdtrakul W, Castro DJ, Gu S, Morry J, Reda M, Gray JW, et al. Current development of targeted oligonucleotide-based cancer therapies: perspective on HER2-positive breast cancer treatment. Cancer Treat Rev. 2016;45:19–29.
Ma L, Liang Z, Zhou H, Qu L. Applications of RNA indexes for precision oncology in breast cancer. Genom Proteom Bioinform. 2018;16(2):108–19.
Bramsen JB, Laursen MB, Nielsen AF, Hansen TB, Bus C, Langkjaer N, et al. A large-scale chemical modification screen identifies design rules to generate siRNAs with high activity, high stability and low toxicity. Nucleic Acids Res. 2009;37(9):2867–81.
Baumann V, Winkler J. miRNA-based therapies: strategies and delivery platforms for oligonucleotide and non-oligonucleotide agents. Future Med Chem. 2014;6(17):1967–84.
Bader AG. miR-34—a microRNA replacement therapy is headed to the clinic. Front Genet. 2012;3:120.
Anuj LA, Venkatraman G Rayala SK. Increased expression of MicroRNA 551a by c-Fos reduces focal adhesion kinase levels and blocks tumorigenesis. Mol Cell Biol 2019;39(7):e00577-18.
Kong X, Li G, Yuan Y, He Y, Wu X, Zhang W, et al. MicroRNA-7 inhibits epithelial-to-mesenchymal transition and metastasis of breast cancer cells via targeting FAK expression. PLoS ONE. 2012;7(8): e41523.
Teo MT, Landi D, Taylor CF, Elliott F, Vaslin L, Cox DG, et al. The role of microRNA-binding site polymorphisms in DNA repair genes as risk factors for bladder cancer and breast cancer and their impact on radiotherapy outcomes. Carcinogenesis. 2012;33(3):581–6.
Hannafon BN, Cai A, Calloway CL, Xu Y-F, Zhang R, Fung K-M, et al. miR-23b and miR-27b are oncogenic microRNAs in breast cancer: evidence from a CRISPR/Cas9 deletion study. BMC Cancer. 2019;19(1):642.
Li W, Li G, Fan Z, Liu T. Tumor-suppressive microRNA-452 inhibits migration and invasion of breast cancer cells by directly targeting RAB11A. Oncol Lett. 2017;14(2):2559–65.
Han M, Hu J, Lu P, Cao H, Yu C, Li X, et al. Exosome-transmitted miR-567 reverses trastuzumab resistance by inhibiting ATG5 in breast cancer. Cell Death Dis. 2020;11(1):43.
Ao X, Nie P, Wu B, Xu W, Zhang T, Wang S, et al. Decreased expression of microRNA-17 and microRNA-20b promotes breast cancer resistance to taxol therapy by upregulation of NCOA3. Cell Death Dis. 2016;7(11):e2463–e2463.
Lou W, Liu J, Ding B, Xu L, Fan W. Identification of chemoresistance-associated miRNAs in breast cancer. Cancer Manag Res. 2018;10:4747–57.
Xue J, Chi Y, Chen Y, Huang S, Ye X, Niu J, et al. MiRNA-621 sensitizes breast cancer to chemotherapy by suppressing FBXO11 and enhancing p53 activity. Oncogene. 2016;35(4):448–58.
Dettori D, Orso F, Penna E, Baruffaldi D, Brundu S, Maione F, et al. Therapeutic silencing of miR-214 inhibits tumor progression in multiple mouse models. Mol Ther. 2018;26(8):2008–18.
De Cola A, Lamolinara A, Lanuti P, Rossi C, Iezzi M, Marchisio M, et al. MiR-205-5p inhibition by locked nucleic acids impairs metastatic potential of breast cancer cells. Cell Death Dis. 2018;9(8):821.
Wang W, Liu Y, Guo J, He H, Mi X, Chen C, et al. miR-100 maintains phenotype of tumor-associated macrophages by targeting mTOR to promote tumor metastasis via Stat5a/IL-1ra pathway in mouse breast cancer. Oncogenesis. 2018;7(12):97.
Xu J, Sun J, Ho PY, Luo Z, Ma W, Zhao W, et al. Creatine based polymer for codelivery of bioengineered MicroRNA and chemodrugs against breast cancer lung metastasis. Biomaterials. 2019;210:25–40.
Huang Z, Gan J, Long Z, Guo G, Shi X, Wang C, et al. Targeted delivery of let-7b to reprogramme tumor-associated macrophages and tumor infiltrating dendritic cells for tumor rejection. Biomaterials. 2016;90:72–84.
Zhang Q, Ran R, Zhang L, Liu Y, Mei L, Zhang Z, et al. Simultaneous delivery of therapeutic antagomirs with paclitaxel for the management of metastatic tumors by a pH-responsive anti-microbial peptide-mediated liposomal delivery system. J Control Release. 2015;197:208–18.
Li L, Xie X, Luo J, Liu M, Xi S, Guo J, et al. Targeted expression of miR-34a using the T-VISA system suppresses breast cancer cell growth and invasion. Mol Ther. 2012;20(12):2326–34.
Yin H, Xiong G, Guo S, Xu C, Xu R, Guo P, et al. Delivery of Anti-miRNA for triple-negative breast cancer therapy using RNA nanoparticles targeting stem cell marker CD133. Mol Ther. 2019;27(7):1252–61.
Deng X, Cao M, Zhang J, Hu K, Yin Z, Zhou Z, et al. Hyaluronic acid-chitosan nanoparticles for co-delivery of MiR-34a and doxorubicin in therapy against triple negative breast cancer. Biomaterials. 2014;35(14):4333–44.
Ramchandani D, Lee SK, Yomtoubian S, Han MS, Tung C-H, Mittal V. Nanoparticle delivery of miR-708 mimetic impairs breast cancer metastasis. Mol Cancer Ther. 2019;18(3):579–91.
Acknowledgements
We acknowledge the Central University of Punjab for providing infrastructure and facilities. Council of Scientific and Industrial Research (CSIR), New Delhi, for Senior Research Fellowship (SRF) to Yogita Chhichholiya and Science and Engineering Research Board (SERB) to Harsh Vikram Singh for Project Assistantship. DST-FIST grant (SR/FST/LS-I/2017/49) to the Department of Human Genetics and Molecular Medicine, Central University of Punjab, is acknowledged with thanks.
Funding
Financial support to Yogita Chhichholiya (Award No- 09/1051(0038)/2019-EMR-1) from Council of Scientific and Industrial Research (CSIR) and Harsh Vikram Singh (Sanction No-SRG/2021/001806) from Science and Engineering Research Board (SERB) India is highly acknowledged.
Author information
Authors and Affiliations
Contributions
AM, SS and YC conceptualized the idea of the review. YC and HVS curated the data and prepared the draft. AM critically revised and edited the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Chhichholiya, Y., Singh, H.V., Singh, S. et al. Genetic variations in tumor-suppressor miRNA-encoding genes and their target genes: focus on breast cancer development and possible therapeutic strategies. Clin Transl Oncol 26, 1–15 (2024). https://doi.org/10.1007/s12094-023-03176-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12094-023-03176-8