Abstract
Background
Acute myeloid leukemia (AML) is a highly heterogeneous hematological cancer. The current diagnosis and therapy model of AML has gradually shifted to personalization and accuracy. Artesunate, a member of the artemisinin family, has anti-tumor impacts on AML. This research uses network pharmacology and molecular docking to anticipate artesunate potential mechanisms of action in the therapy of AML.
Methods
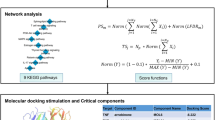
Screening the action targets of artesunate through Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP), PubChem, and Swiss Target Prediction databases; The databases of Online Mendelian Inheritance in Man (OMIM), Disgenet, GeneCards, and Drugbank were utilized to identify target genes of AML, and an effective target of artesunate for AML treatment was obtained through cross-analysis. Protein–protein interaction (PPI) networks are built on the Cytoscape platform. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were conducted on the relevant targets using R software. Finally, using molecular docking technology and Pymol, we performed verification of the effects of active components and essential targets.
Results
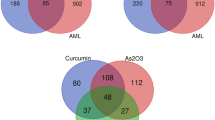
Artesunate 30 effective targets for treating AML include CASP3, EGFR, MAPK1, and STAT3, four targeted genes that may have a crucial function in disease management. The virus infection-related pathway (HeptatisB (HBV), Human papillomavirus (HPV), Epstein-Barr virus (EBV) infection and etc.), FoxO, viral carcinogenesis, and proteoglycans in cancer signaling pathways have all been hypothesized to be involved in the action mechanism of GO, which is enriched in 2044 biological processes, 125 molecular functions, 209 cellular components, and 106 KEGG pathways. Molecular docking findings revealed that artesunate was critically important in the therapy of AML due to its high affinity for the four primary disease targets. Molecular docking with a low binding energy yields helpful information for developing medicines against AML.
Conclusions
Consequently, artesunate may play a role in multi-targeted, multi-signaling pathways in treating AML, suggesting that artesunate may have therapeutic potential for AML.






Similar content being viewed by others
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
References
Döhner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med. 2015;373:1136–52.
Dong Y, Shi O, Zeng Q, Lu X, Wang W, Li Y, et al. Leukemia incidence trends at the global, regional, and national level between 1990 and 2017. Exp Hematol Oncol [Internet]. 2020 [cited 2022 Dec 11]; 9:14. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7304189/
Rowe JM. Perspectives on current survival and new developments in AML. Best Pract Res Clin Haematol [Internet]. 2021;34:101248.
Kantarjian H, Kadia T, DiNardo C, Daver N, Borthakur G, Jabbour E, et al. Acute myeloid leukemia: current progress and future directions. Blood Cancer J. 2021;11:41.
Döhner H, Wei AH, Löwenberg B. Towards precision medicine for AML. Nat Rev Clin Oncol. 2021;18:577–90.
Wang J, Wong Y-K, Liao F. What has traditional Chinese medicine delivered for modern medicine? Expert Rev Mol Med. 2018;20:e4.
Wang J, Xu C, Wong YK, Liao FL, Jiang T, Tu Y. Malaria eradication. Lancet. 2020;395:e69.
Tu Y. The discovery of artemisinin (qinghaosu) and gifts from Chinese medicine. Nat Med. 2011;17:1217–20.
Tu Y. Artemisinin-A gift from traditional chinese medicine to the world (nobel lecture). Angew Chem Int Ed Engl. 2016;55:10210–26.
Ma N, Zhang Z, Liao F, Jiang T, Tu Y. The birth of artemisinin. Pharmacol Ther. 2020;216:107658.
Zhang J, Wang Y, Yin C, Gong P, Zhang Z, Zhao L, et al. Artesunate improves venetoclax plus cytarabine AML cell targeting by regulating the Noxa/Bim/Mcl-1/p-Chk1 axis. Cell Death Dis. 2022;13:379.
Nogales C, Mamdouh ZM, List M, Kiel C, Casas AI, Schmidt HHHW. Network pharmacology: curing causal mechanisms instead of treating symptoms. Trends Pharmacol Sci [Internet]. 2022;43:136–50.
Pinzi L, Rastelli G. Molecular docking: shifting paradigms in drug discovery. Int J Mol Sci. 2019;20:4331.
Ru J, Li P, Wang J, Zhou W, Li B, Huang C, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform. 2014;6:13.
Halladay CW, Trikalinos TA, Schmid IT, Schmid CH, Dahabreh IJ. Using data sources beyond PubMed has a modest impact on the results of systematic reviews of therapeutic interventions. J Clin Epidemiol. 2015;68:1076–84.
Daina A, Michielin O, Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019;47:W357–64.
Amberger JS, Hamosh A. Searching online mendelian inheritance in man (OMIM): a knowledgebase of human genes and genetic phenotypes. Curr Protoc Bioinformatics. 2017;58:121–1212.
Piñero J, Bravo À, Queralt-Rosinach N, Gutiérrez-Sacristán A, Deu-Pons J, Centeno E, et al. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2017;45:D833–9.
Safran M, Chalifa-Caspi V, Shmueli O, Olender T, Lapidot M, Rosen N, et al. Human gene-centric databases at the weizmann institute of science: GeneCards, UDB, CroW 21 and HORDE. Nucleic Acids Res. 2003;31:142–6.
Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10:1523.
Valente TW, Fujimoto K. Bridging: locating critical connectors in a network. Soc Networks. 2010;32:212–20.
Sahin Y. LncRNA H19 is a potential biomarker and correlated with immune infiltration in thyroid carcinoma. Clin Exp Med. 2022. https://doi.org/10.1007/s10238-022-00853-w.
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, et al. The protein data bank. Nucleic Acids Res. 2000;28:235–42.
Ma Z, Woon CY-N, Liu C-G, Cheng J-T, You M, Sethi G, et al. Repurposing artemisinin and its derivatives as anticancer drugs: a chance or challenge? Front Pharmacol. 2021;12:828856.
Abba ML, Patil N, Leupold JH, Saeed MEM, Efferth T, Allgayer H. Prevention of carcinogenesis and metastasis by Artemisinin-type drugs. Cancer Lett. 2018;429:11–8.
Efferth T. Cancer combination therapies with artemisinin-type drugs. Biochem Pharmacol. 2017;139:56–70.
Mancuso RI, Foglio MA, Olalla Saad ST. Artemisinin-type drugs for the treatment of hematological malignancies. Cancer Chemother Pharmacol. 2021;87:1–22.
Lu X, Efferth T. Repurposing of artemisinin-type drugs for the treatment of acute leukemia. Semin Cancer Biol. 2021;68:291–312.
Zhao F, Vakhrusheva O, Markowitsch SD, Slade KS, Tsaur I, Cinatl J, et al. Artesunate impairs growth in cisplatin-resistant bladder cancer cells by cell cycle arrest, apoptosis and autophagy induction. Cells [Internet]. 2020;9:2643.
Estrov Z, Thall PF, Talpaz M, Estey EH, Kantarjian HM, Andreeff M, et al. Caspase 2 and caspase 3 protein levels as predictors of survival in acute myelogenous leukemia. Blood. 1998;92:3090–7.
Shalini S, Dorstyn L, Dawar S, Kumar S. Old, new and emerging functions of caspases. Cell Death Differ. 2015;22:526–39.
Huang Q, Li F, Liu X, Li W, Shi W, Liu F-F, et al. Caspase 3-mediated stimulation of tumor cell repopulation during cancer radiotherapy. Nat Med [Internet]. 2011;17:860–6.
Carlile GW, Smith DH, Wiedmann M. Caspase-3 has a nonapoptotic function in erythroid maturation. Blood. 2004;103:4310–6.
Man N, Tan Y, Sun X-J, Liu F, Cheng G, Greenblatt SM, et al. Caspase-3 controls AML1-ETO-driven leukemogenesis via autophagy modulation in a ULK1-dependent manner. Blood. 2017;129:2782–92.
Cai S, Zhang Y-X, Han K, Ding Y-Q. Expressions and clinical significance of COX-2, VEGF-C, and EFGR in endometrial carcinoma. Arch Gynecol Obstet. 2017;296:93–8.
Thomas NJ, Myall NJ, Sun F, Patil T, Mushtaq R, Yu C, et al. Brain metastases in EGFR- and ALK-positive NSCLC: outcomes of central nervous system-penetrant tyrosine kinase inhibitors alone versus in combination with radiation. J Thorac Oncol. 2022;17:116–29.
Gehart H, Kumpf S, Ittner A, Ricci R. MAPK signalling in cellular metabolism: stress or wellness? EMBO Rep [Internet]. 2010;11:834–40.
D’Souza WN, Chang C-F, Fischer AM, Li M, Hedrick SM. The Erk2 MAPK regulates CD8 T cell proliferation and survival. J Immunol. 2008;181:7617–29.
Moshofsky KB, Cho HJ, Wu G, Romine KA, Newman MT, Kosaka Y, et al. Acute myeloid leukemia–induced T-cell suppression can be reversed by inhibition of the MAPK pathway. Blood Adv [Internet]. 2019;3:3038–51.
Turkson J. STAT proteins as novel targets for cancer drug discovery. Expert Opin Ther Targets. 2004;8:409–22.
Zou S, Tong Q, Liu B, Huang W, Tian Y, Fu X. Targeting STAT3 in cancer immunotherapy. Mol Cancer. 2020;19:145.
Amaya ML, Inguva A, Pei S, Jones C, Krug A, Ye H, et al. The STAT3-MYC axis promotes survival of leukemia stem cells by regulating SLC1A5 and oxidative phosphorylation. Blood. 2022;139:584–96.
Rezvani K, Barrett J. STAT3: the “Achilles” heel for AML? Blood. 2014;123:1–2.
Guo Y, Wang W, Sun H. A systematic review and meta-analysis on the risk factors of acute myeloid leukemia. Transl Cancer Res. 2022;11:796–804.
Movassagh M, Oduor C, Forconi C, Moormann AM, Bailey JA. Sensitive detection of EBV microRNAs across cancer spectrum reveals association with decreased survival in adult acute myelocytic leukemia. Sci Rep. 2019;9:20321.
Chen C-Y, Huang S-Y, Cheng A, Chou W-C, Yao M, Tang J-L, et al. High risk of hepatitis b reactivation among patients with acute myeloid leukemia. PLoS ONE. 2015;10:e0126037.
Farhan M, Wang H, Gaur U, Little PJ, Xu J, Zheng W. FOXO signaling pathways as therapeutic targets in cancer. Int J Biol Sci [Internet]. 2017;13:815–27.
Long J, Jia M-Y, Fang W-Y, Chen X-J, Mu L-L, Wang Z-Y, et al. FLT3 inhibition upregulates HDAC8 via FOXO to inactivate p53 and promote maintenance of FLT3-ITD+ acute myeloid leukemia. Blood [Internet]. 2020;135:1472–83.
Funding
This work was supported by the National Natural Science Foundation of China (No.81873286), Program of Shanghai Academic Research Leader (No.20XD1403500), Shanghai Science Technology and Innovation Action Plan (No.21Y31920400), Clinical Science and Technology Innovation Project of Shanghai Shenkang Hospital Development Center (No. SHDC12020128).
Author information
Authors and Affiliations
Contributions
All authors contributed to the article and approved the submitted version. YT and WL participated in data collection and analysis. JY, TX, YW, XD, HX, and JR participated in manuscript writing. JL is responsible for the integrity and accuracy of the data.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ethical approval and informed consent
We declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tao, Y., Li, W., Yang, J. et al. Exploring underlying mechanism of artesunate in treatment of acute myeloid leukemia using network pharmacology and molecular docking. Clin Transl Oncol 25, 2427–2437 (2023). https://doi.org/10.1007/s12094-023-03125-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12094-023-03125-5