Abstract
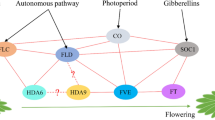
Histone acetylation is an important posttranslational modification associated with gene activation. In Arabidopsis, histone acetyltransferase 1 (HAC1) can promote flowering by regulating the transcription of FLOWERING LOCUS C (FLC), a major floral repressor. The size of the full-length cDNA and genomic DNA sequences of the histone acetyltransferase 1 gene (BrcuHAC1) in Chinese flowering cabbage (Brassica rapa syn. campestris ssp. chinensis var. utilis) were 5846 bp and 7376 bp, with an open reading frame (ORF) coding for a peptide with 1689 amino acids. The expression levels of BrcuHAC1 in different tissues and different developmental stages were as follows: flower > leaf > stem > root, and completed bolting and flowering stage > 5th true leaf-stage > 4th true leaf-stage > 3rd true leaf-stage > 2nd true leaf-stage > 1st true leaf-stage. Silencing of BrcuHAC1 resulted in slow growth, and delayed bolting and flowering time in Chinese flowering cabbage. Molecular analysis showed that the mRNA level of FLC was increased, indicating that the delayed flowering phenomenon was mediated by FLC in the silenced group. In contrast, the expression levels of the autonomous-pathway genes were not significantly affected in the silenced group. In addition, the histone modification of FLC chromatin was also not affected in the silenced group. FLC is not the direct target gene of BrcuHAC1. However, BrcuHAC1 may affect the bolting and flowering time of Chinese flowering cabbage through the epigenetic modification of upstream factors of FLC.






Similar content being viewed by others
References
Ausin I., Alonso-Blanco C., Jarillo J. A., Ruiz-Garcia L. and Martinez-Zapater J. M. 2004 Regulation of flowering time by FVE, a a retinoblastoma-associated protein. Nat. Genet. 36, 162–166.
Benhamed M., Bertrand C., Servet C. and Zhou D. X. 2006 Arabidopsis GCN5, HD1, and TAF1/HAF2 interact to regulate histone acetylation required for light responsive gene expression. Plant Cell. 18, 2893–2903.
Bereshchenko O. R., Gu W. and Dalla-Favera R. 2002 Acetylation inactivates the transcriptional repressor BCL6. Nat. Genet. 32, 606–613.
Berge S. L. 2007 The complex language of chromatin regulation during transcription. Nature 447, 407–412.
Bharti K., Koskull-Döring P. V., Bharti S., Kumar P., Tintschl-Korbitzer A., Treuter E. and Nover L. 2004 Tomato heat stress transcription factor HsfB1 represents a novel type of general transcription coactivator with a histone-like motif interacting with the plant CREB binding protein ortholog HAC1. Plant Cell 16, 1521–1535.
Bienz M. 2006 The PHD finger, a nuclear protein-interaction domain. Trends Biochem. Sci. 31, 35–40.
Bordoli L., Netsch M., Lüthi U., Lutz W. and Eckner R. 2001 Plant orthologs of p300/CBP: conservation of a core domain in metazoan p300/CBP acetyltransferase-related proteins. Nucleic Acids Res. 29, 589–597.
Boycheva I., Vassileva V. and Iantcheva A. 2014 Histone acetyltransferases in plant development and plasticity. Curr. Genomics 15, 28–37.
Chen Z., Zhang H., Jablonowski D., Zhou X., Ren X., Hong X. et al. 2006 Mutations in ABO1/ELO2, a subunit of holo-Elongator, increase abscisic acid sensitivity and drought tolerance in Arabidopsis thaliana. Mol. Cell. Biol. 26, 6902–6912.
Chung E., Seong E., Kim Y. C., Chung E. J., Oh S. K., Lee S. et al. 2004 A method of high frequency virus-induced gene silencing in chili pepper (Capsicum annuum L. cv. Bukang). Mol. Cells 17, 377–380.
Deng W. W., Liu C., Pei Y., Deng X., Niu L. and Cao X. 2007 Involvement of the histone acetyltransferase AtHAC1 in the regulation of flowering time via repression of FLOWERING LOCUS C in Arabidopsis. Plant Physiol. 143, 1660–1668.
Fisher A. J. and Franklin K. A. 2011 Chromatin remodeling in plant light signaling. Physiol. Plant. 142, 305–313.
Fornara F., Montaigu A. and Coupland G. SnapShot: Control of flowering in Arabidopsis. Cell 141, 550 e1–e2.
Gamsjaeger R., Liew C. K., Loughlin F. E., Crossley M. and Mackay J. P. 2007 Sticky fingers: zinc fingers as protein-recognition motifs. Trends. Biochem. Sci. 32, 63–70.
Guidez F., Howell L. and Isalan M. 2005 Histone acetyltransferase activity of p300 is required for transcriptional repression by the promyeolocytic leukemia zinc finger protein. Mol. Cell. Biol. 25, 5552–5566.
Guo L., Zhou J., Elling A. A., Charron J. B. and Deng X. W. 2008 Histone modifications and expression of light-regulated genes in Arabidopsis are cooperatively influenced by changing light conditions. Plant Physiol. 147, 2070–2083.
Han S. K., Song J. D., Noh Y. S. and Noh B. 2007 Role of plant CBP/p300-like genes in the regulation of flowering time. Plant J. 49, 103–114.
He Y. 2009 Control of the transition to flowering by chromatin modifications. Mol. Plant 2, 554–564.
Hinckley W. E., Keymanesh K., Cordova J. A. and Brusslan J. A. 2019 The HAC1 histone acetyltransferase promotes leaf senescence and regulates the expression of ERF022. Plant Direct 3, e00159.
Jin H., Choi S. M., Kang M. J., Yun S. H., Kwon D. J., Noh Y. S. and Noh B. 2018 Salicylic acid-induced transcriptional reprogramming by the HAC-NPR1-TGA histone acetyltransferase complex in Arabidopsis. Nucleic Acids Res. 46, 11712–11725.
Kalkhoven E. 2004 CBP and p300: HATs for different occasions. Biochem. Pharmacol. 68, 1145–1155.
Kawasaki H., Eckner R., Yao T. P., Taira K., Chiu R., Livingston D. M. and Yokoyama K. K. 2004 Distinct roles of the co-activators p300 and CBP in retinoic-acid-induced F9-cell differentiation. Nature 393, 284–289.
Korzus E., Rosenfeld M. G. and Mayford M. 2004 CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron 42, 961–972.
Kovar H. 2011 Dr. Jekyll and Mr. Hyde: the Two Faces of the FUS/EWS/TAF15 Protein Family. Sarcoma 2011, 1–13.
Lallous N., Legrand P., McEwen A. G., Ramon-Maiques S., Samama J. P. and Birck C. 2011 The PHD finger of human UHRF1 reveals a new subgroup of unmethylated histone H3 tail readers. PLoS One 6, e27599.
Li C., Xu J., Li J., Li Q. and Yang H. 2014a Involvement of Arabidopsis HAC family genes in pleiotropic developmental processes. Plant. Signal. Behav. 9, e28173.
Li C., Xu J., Li J., Li Q. and Yang H. 2014b Involvement of Arabidopsis histone acetyltransferase HAC family genes in the ethylene signaling pathway. Plant Cell Physiol. 55, 426–435.
Liu Y., Nakayama N., Schiff M., Litt A., Irish V. F. and Dinesh-Kumar S. P. 2009 Virus-induced gene silencing of a DEFICIENS ortholog in Nicotiana benthamiana. Plant. Mol. Biol. 54, 701–711.
Liu Y., Schiff M. and Dinesh-Kumar S. P. 2002 Virus–induced gene silencing in tomato. Plant J. 31, 777–786.
Michaels S. D. and Amasino R. M. 1999 FLOWEING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11, 949–956.
Michaels S. D. and Amasino R. M. 2001 The main flowering pathways in A. thaliana, defined as the autonomous and vernalization, inhibit FLC expression. Plant Cell 13, 935–941.
Michaels S. D., Bezerra I. C. and Amasino R. M. 2004 FRIGIDA-related genes are required for the winter-annual habit in Arabidopsis. Proc. Natl. Acad. Sci. USA 101, 3281–3285.
Nakamura M. and Hennig L. 2017 Inheritance of vernalization memory at FLOWERING LOCUS C during plant regeneration. J. Exp. Bot. 68, 2813–2819.
Neuwald A. F. and Landsman D. 1997 GCN5–related histone N–acetyltransferases belong to a diverse superfamily that includes the yeast SPT10 protein. Trends. Biochem. Sci. 22, 154–155.
Pandey R., Muller A., Napoli C. A., Selinger D. A., Pikaard C. S., Richards E. J., Bender J., Mount D. W. and Jorgensen R. A. 2002 Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 30, 5036–5055.
Pavangadkar K., Thomashow M. F. and Triezenberg S. J. 2010 Histone dynamics and roles of histone acetyltransferases during cold-induced gene regulation in Arabidopsis. Plant. Mol. Biol. 74, 183–200.
Salathia N., Davis S. J., Lynn J. R., Michaels S. D., Amasino R. M. and Millar A. J. 2006 FLOWERING LOCUS C-dependent and -independent regulation of the circadian clock by the autonomous and vernalization pathways. BMC Plant. Biol. 6, 10.
Schwartz J. C., Cech T. R. and Parker R. R. 2015 Biochemical properties and biological functions of FET proteins. Annu. Rev. Biochem. 84, 355–379.
Servet C., Conde E., Silva N. and Zhou D. X. 2010 Histone acetyltransferase AtGCN5/HAG1 is a versatile regulator of developmental and inducible gene expression in Arabidopsis. Mol. Plant 3, 670–677.
Sokol A., Kwiatkowska A., Jerzmanowski A. and Prymakowska-Bosak M. 2007 Upregulation of stress-inducible genes in tobacco and Arabidopsis cells in response to abiotic stresses and ABA treatment correlates with dynamic changes in histone H3 and H4 modifications. Planta 227, 245–254.
Stockinger E. J., Mao Y., Regier M. K., Triezenberg S. J. and Thomashow M. F. 2001 Transcriptional adaptor and histone acetyltransferase proteins in Arabidopsis and their interactions with CBF1, a transcriptional activator involved in cold-regulated gene expression. Nucleic Acids Res. 29, 1524–1533.
Tian L., Fong M. P., Wang J. J., Wei J. Y., Jiang H. M., Doerge R. W. and Chen Z. J. 2005 Reversible histone acetylation and deacetylation mediate genome-wide, promoter-dependent and locus-specific changes in gene expression during plant development. Genetics 169, 337–345.
Vlachonasios K. E., Thomashow M. F. and Triezenberg S. J. 2003 Disruption mutations of ADA2b and GCN5 transcriptional adaptor genes dramatically affect Arabidopsis growth, development, and gene expression. Plant Cell. 15, 626–638.
Whittaker C. and Dean C. 2017 The FLC locus: a platform for discoveries in epigenetics and adaptation. Annu. Rev. Cell. Dev. Biol. 33, 555–575.
Xiao X. F., Cao B. H., Wang Y., Chen G. J. and Lei J. J. 2008 Study on flower bud differentiation and cloning and expression of BrcuFLC in Brassica campestris L. ssp. chinensis (L.) Makino var. utilis. Acta. Horti. Sinca. 35, 827–832.
Xiao X. F., Lei J. J., Cao B. H., Chen G. J. and Chen C. M. 2012 cDNA-AFLP analysis on bolting or flowering of Chinese flowering cabbage and molecular characteristics of BrcuDFR–like/BrcuAXS gene. Mol. Biol. Rep. 39, 7525–7531.
Xing L. J., Liu Y., Xu S. J., Xiao J., Wang B., Deng H. W. 2018 Arabidopsis O‐GlcNAc transferase SEC activates histone methyltransferase ATX1 to regulate flowering. EMBO J. 37, e98115.
Yuan Z. L., Guan Y. J., Chatterjee D. and Chin Y. E. 2005 Stat3 dimerization regulated by reversible acetylation of single lysine residue. Science 307, 269–273.
Zhou X., Hua D., Chen Z., Zhou Z. and Gong Z. 2009 Elongator mediates ABA responses, oxidative stress resistance and anthocyanin biosynthesis in Arabidopsis. Plant J. 60, 79–90.
Acknowledgement
This research was funded by the National Natural Science Foundation of China (grant no. 31860560 and 31360484).
Author information
Authors and Affiliations
Corresponding author
Additional information
Corresponding editor: Manoj Prasad
Shucheng Si and Ming Zhang contributed equally to this work.
Rights and permissions
About this article
Cite this article
Si, S., Zhang, M., Hu, Y. et al. BrcuHAC1 is a histone acetyltransferase that affects bolting development in Chinese flowering cabbage. J Genet 100, 56 (2021). https://doi.org/10.1007/s12041-021-01303-4
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12041-021-01303-4