Abstract
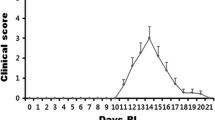
Experimental autoimmune encephalomyelitis (EAE) is an animal model of multiple sclerosis that shows demyelination in the central nervous system and functional deficits, including olfactory impairment. However, the genes related to olfactory impairment in EAE are unknown. We evaluated hub genes of the olfactory bulb in EAE mice. Differentially expressed genes (cut-offs, fold change > 2 and adjusted p < 0.05) and their related pathways in olfactory bulbs were subjected to gene ontology (GO) pathway analysis, gene set enrichment analysis (GSEA). Protein-protein interactions with selected genes were evaluated using the Search Tool for the Retrieval of Interacting Genes/Proteins. Gene regulatory networks (GRNs) which were constructed at the post-transcriptional level, including the genes-transcription factors (TFs) and gene-microRNAs (miRNAs) interaction networks. Twelve hub genes were found, three of which (Ctss, Itgb2, and Tlr2) were validated by RT-qPCR to be related to GO pathways such as immune response and regulation of immune response. GSEA showed that neuron-related genes—including Atp6v1g2, Egr1, and Gap43—and their pathways were significantly downregulated. GRNs analysis of six genes (Ctss, Itgb2, Tlr2, Atp6v1g2, Egr1, and Gap43) revealed 37 TFs and 84 miRNAs were identified as potential regulators of six genes, indicating significant interaction among six genes, TFs, and miRNAs. Collectively, these results suggest that transcriptomic analysis of the olfactory bulb of EAE mice can provide insight into olfactory dysfunction and reveal therapeutic targets for olfactory impairment.











Similar content being viewed by others
Data Availability
Data will be made available on request.
Abbreviations
- BP:
-
Biological process
- CC:
-
Cellular component
- cDNA:
-
Complementary deoxyribonucleic acid
- CNS:
-
Central nervous system
- DEG:
-
Differentially expressed gene
- EAE:
-
Experimental autoimmune encephalomyelitis
- FDR:
-
False discovery rate
- GAPDH:
-
Glyceraldehyde 3-phosphate dehydrogenase
- GO:
-
Gene ontology
- GSEA:
-
Gene set enrichment analysis
- Iba1:
-
Ionized calcium-binding adapter molecule 1
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- MCC:
-
Maximal clique centrality
- MF:
-
Molecular function
- MHC:
-
Major histocompatibility complex
- MNC:
-
Maximum neighborhood component
- MOG:
-
Myelin oligodendrocyte glycoprotein
- MS:
-
Multiple sclerosis
- MSigDB:
-
Molecular signatures database
- NOM:
-
Nominal
- PC:
-
Principal component
- PCA:
-
Principal component analysis
References
Shin T, Ahn M, Matsumoto Y (2012) Mechanism of experimental autoimmune encephalomyelitis in Lewis rats: recent insights from macrophages. Anat Cell Biol 45(3):141–148. https://doi.org/10.5115/acb.2012.45.3.141
Shin T, Kojima T, Tanuma N, Ishihara Y, Matsumoto Y (1995) The subarachnoid space as a site for precursor T cell proliferation and effector T cell selection in experimental autoimmune encephalomyelitis. J Neuroimmunol 56(2):171–178. https://doi.org/10.1016/0165-5728(94)00144-d
Shrestha B, Jiang X, Ge S, Paul D, Chianchiano P, Pachter JS (2017) Spatiotemporal resolution of spinal meningeal and parenchymal inflammation during experimental autoimmune encephalomyelitis. Neurobiol Dis 108:159–172. https://doi.org/10.1016/j.nbd.2017.08.010
Villarroya H, Violleau K, Ben Younes-Chennoufi A, Baumann N (1996) Myelin-induced experimental allergic encephalomyelitis in Lewis rats: tumor necrosis factor alpha levels in serum and cerebrospinal fluid immunohistochemical expression in glial cells and macrophages of optic nerve and spinal cord. J Neuroimmunol 64(1):55–61. https://doi.org/10.1016/0165-5728(95)00151-4
Goektas O, Schmidt F, Bohner G, Erb K, Ludemann L, Dahlslett B, Harms L, Fleiner F (2011) Olfactory bulb volume and olfactory function in patients with multiple sclerosis. Rhinology 49(2):221–226. https://doi.org/10.4193/Rhino10.136
Caminiti F, De Salvo S, De Cola MC, Russo M, Bramanti P, Marino S, Ciurleo R (2014) Detection of olfactory dysfunction using olfactory event related potentials in young patients with multiple sclerosis. PLoS One 9(7):e103151. https://doi.org/10.1371/journal.pone.0103151
Kapadia M, Stanojcic M, Earls AM, Pulapaka S, Lee J, Sakic B (2012) Altered olfactory function in the MRL model of CNS lupus. Behav Brain Res 234(2):303–311. https://doi.org/10.1016/j.bbr.2012.07.005
Gaillard I, Rouquier S, Giorgi D (2004) Olfactory receptors. Cell Mol Life Sci 61(4):456–469. https://doi.org/10.1007/s00018-003-3273-7
Kovács T (2004) Mechanisms of olfactory dysfunction in aging and neurodegenerative disorders. Ageing Res Rev 3(2):215–232. https://doi.org/10.1016/j.arr.2003.10.003
Kim J, Ahn M, Choi Y, Shin T (2020) Upregulation of cathepsins in olfactory bulbs is associated with transient olfactory dysfunction in mice with experimental autoimmune encephalomyelitis. Molecular neurobiology 57(8):3412–3423. https://doi.org/10.1007/s12035-020-01952-z
Kim J, Ahn M, Choi Y, Ekanayake P, Park CM, Moon C, Jung K, Tanaka A, Matsuda H, Shin T (2019) Gene expression profile of olfactory transduction signaling in an animal model of human multiple sclerosis. Exp Neurobiol 28(1):74–84. https://doi.org/10.5607/en.2019.28.1.74
Kim J, Choi Y, Ahn M, Jung K, Shin T (2018) Olfactory dysfunction in autoimmune central nervous system neuroinflammation. Molecular neurobiology 55(11):8499–8508. https://doi.org/10.1007/s12035-018-1001-4
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9(4):357–359. https://doi.org/10.1038/nmeth.1923
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15(12):550. https://doi.org/10.1186/s13059-014-0550-8
Team RC (2013) R: a language and environment for statistical computing.
Ge SX, Jung D, Yao R (2020) ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics 36(8):2628–2629. https://doi.org/10.1093/bioinformatics/btz931
Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, Doncheva NT, Legeay M, Fang T, Bork P, Jensen LJ, von Mering C (2021) The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res 49(D1):D605–D612. https://doi.org/10.1093/nar/gkaa1074
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY (2014) cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol 8:S11. https://doi.org/10.1186/1752-0509-8-s4-s11
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13(11):2498–2504. https://doi.org/10.1101/gr.1239303
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102(43):15545–15550. https://doi.org/10.1073/pnas.0506580102
Zhou G, Soufan O, Ewald J, Hancock REW, Basu N, Xia J (2019) NetworkAnalyst 3.0: a visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res 47(W1):W234–w241. https://doi.org/10.1093/nar/gkz240
Karagkouni D, Paraskevopoulou MD, Chatzopoulos S, Vlachos IS, Tastsoglou S, Kanellos I, Papadimitriou D, Kavakiotis I, Maniou S, Skoufos G, Vergoulis T, Dalamagas T, Hatzigeorgiou AG (2018) DIANA-TarBase v8: a decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res 46(D1):D239–d245. https://doi.org/10.1093/nar/gkx1141
Chen Y, Wang X (2020) miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res 48(D1):D127–d131. https://doi.org/10.1093/nar/gkz757
Janky R, Verfaillie A, Imrichová H, Van de Sande B, Standaert L, Christiaens V, Hulselmans G, Herten K, Naval Sanchez M, Potier D, Svetlichnyy D, Kalender Atak Z, Fiers M, Marine JC, Aerts S (2014) iRegulon: from a gene list to a gene regulatory network using large motif and track collections. PLoS Comput Biol 10(7):e1003731. https://doi.org/10.1371/journal.pcbi.1003731
Consortium EP (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489(7414):57
Reichardt HM, Lühder F (2012) The ambivalent role of apoptosis in experimental autoimmune encephalomyelitis and multiple sclerosis. Curr Pharm Des 18(29):4453–4464. https://doi.org/10.2174/138161212802502224
Tepavčević V, Lazarini F, Alfaro-Cervello C, Kerninon C, Yoshikawa K, Garcia-Verdugo JM, Lledo PM, Nait-Oumesmar B, Baron-Van Evercooren A (2011) Inflammation-induced subventricular zone dysfunction leads to olfactory deficits in a targeted mouse model of multiple sclerosis. J Clin Invest 121(12):4722–4734. https://doi.org/10.1172/jci59145
Erkal B, Vural Korkut S (2022) Identification of miRNAs and their potential effects on multiple sclerosis related pathways using ın silico analysis. Mult Scler Relat Disord 59:103642. https://doi.org/10.1016/j.msard.2022.103642
Weerasinghe-Mudiyanselage PDE, Kang S, Kim JS, Kim JC, Kim SH, Wang H, Shin T, Moon C (2022) Transcriptome profiling in the hippocampi of mice with experimental autoimmune encephalomyelitis. Int J Mol Sci 23(23). https://doi.org/10.3390/ijms232314829
Wang C, Zhou W, Su G, Hu J, Yang P (2022) Progranulin Suppressed autoimmune uveitis and autoimmune neuroinflammation by inhibiting Th1/Th17 cells and promoting treg cells and M2 macrophages. Neurol Neuroimmunol Neuroinflamm 9(2). https://doi.org/10.1212/nxi.0000000000001133
Liu SH, Wang YL, Jiang SM, Wan XJ, Yan JH, Liu CF (2022) Identifying the hub gene and immune infiltration of Parkinson’s disease using bioinformatical methods. Brain Res 1785:147879. https://doi.org/10.1016/j.brainres.2022.147879
Liu C, Xu S, Liu Q, Chai H, Luo Y, Li S (2023) Identification of immune cells infiltrating in hippocampus and key genes associated with Alzheimer’s disease. BMC Med Genomics 16(1):53. https://doi.org/10.1186/s12920-023-01458-2
Shippy DC, Ulland TK (2023) Genome-wide identification of murine interferon genes in microglial-mediated neuroinflammation in Alzheimer’s disease. J Neuroimmunol 375:578031. https://doi.org/10.1016/j.jneuroim.2023.578031
Xie Y, Luo X, He H, Tang M (2021) Novel insight into the role of immune dysregulation in amyotrophic lateral sclerosis based on bioinformatic analysis. Front Neurosci 15:657465. https://doi.org/10.3389/fnins.2021.657465
Benowitz LI, Routtenberg A (1997) GAP-43: an intrinsic determinant of neuronal development and plasticity. Trends Neurosci 20(2):84–91. https://doi.org/10.1016/s0166-2236(96)10072-2
Mason JL, Angelastro JM, Ignatova TN, Kukekov VG, Lin G, Greene LA, Goldman JE (2005) ATF5 regulates the proliferation and differentiation of oligodendrocytes. Mol Cell Neurosci 29(3):372–380. https://doi.org/10.1016/j.mcn.2005.03.004
Greene LA, Lee HY, Angelastro JM (2009) The transcription factor ATF5: role in neurodevelopment and neural tumors. J Neurochem 108(1):11–22. https://doi.org/10.1111/j.1471-4159.2008.05749.x
Angelastro JM, Mason JL, Ignatova TN, Kukekov VG, Stengren GB, Goldman JE, Greene LA (2005) Downregulation of activating transcription factor 5 is required for differentiation of neural progenitor cells into astrocytes. The Journal of neuroscience: the official journal of the Society for Neuroscience 25(15):3889–3899. https://doi.org/10.1523/jneurosci.3447-04.2005
Yang T, Zhang Y, Chen L, Thomas ER, Yu W, Cheng B, Li X (2023) The potential roles of ATF family in the treatment of Alzheimer’s disease. Biomed Pharmacother 161:114544. https://doi.org/10.1016/j.biopha.2023.114544
Hernández IH, Torres-Peraza J, Santos-Galindo M, Ramos-Morón E, Fernández-Fernández MR, Pérez-Álvarez MJ, Miranda-Vizuete A, Lucas JJ (2017) The neuroprotective transcription factor ATF5 is decreased and sequestered into polyglutamine inclusions in Huntington’s disease. Acta Neuropathol 134(6):839–850. https://doi.org/10.1007/s00401-017-1770-2
Li WX, Li GH, Tong X, Yang PP, Huang JF, Xu L, Dai SX (2020) Systematic metabolic analysis of potential target, therapeutic drug, diagnostic method and animal model applicability in three neurodegenerative diseases. Aging 12(10):9882–9914. https://doi.org/10.18632/aging.103253
Dos Santos N, Novaes LS, Dragunas G, Rodrigues JR, Brandão W, Camarini R, Peron JPS, Munhoz CD (2019) High dose of dexamethasone protects against EAE-induced motor deficits but impairs learning/memory in C57BL/6 mice. Scientific reports 9(1):6673. https://doi.org/10.1038/s41598-019-43217-3
Becquart P, Vilariño-Güell C, Quandt JA (2020) Enhanced expression of complement and microglial-specific genes prior to clinical progression in the MOG-experimental autoimmune encephalomyelitis model of multiple sclerosis. Brain Res Bull 165:63–69. https://doi.org/10.1016/j.brainresbull.2020.09.010
Cocchiaro P, De Pasquale V, Della Morte R, Tafuri S, Avallone L, Pizard A, Moles A, Pavone LM (2017) The multifaceted role of the lysosomal protease cathepsins in kidney disease. Front Cell Dev Biol 5:114. https://doi.org/10.3389/fcell.2017.00114
Lutzner N, Kalbacher H (2008) Quantifying cathepsin S activity in antigen presenting cells using a novel specific substrate. J Biol Chem 283(52):36185–36194. https://doi.org/10.1074/jbc.M806500200
Baugh M, Black D, Westwood P, Kinghorn E, McGregor K, Bruin J, Hamilton W, Dempster M, Claxton C, Cai J, Bennett J, Long C, McKinnon H, Vink P, den Hoed L, Gorecka M, Vora K, Grant E, Percival MD et al (2011) Therapeutic dosing of an orally active, selective cathepsin S inhibitor suppresses disease in models of autoimmunity. J Autoimmun 36(3-4):201–209. https://doi.org/10.1016/j.jaut.2011.01.003
Arnaout MA (1990) Structure and function of the leukocyte adhesion molecules CD11/CD18. Blood 75(5):1037–1050
Geng XD, Wang WW, Feng Z, Liu R, Cheng XL, Shen WJ, Dong ZY, Cai GY, Chen XM, Hong Q, Wu D (2019) Identification of key genes and pathways in diabetic nephropathy by bioinformatics analysis. J Diabetes Investig 10(4):972–984. https://doi.org/10.1111/jdi.12986
Wasko NJ, Nichols F, Clark RB (2020) Multiple sclerosis, the microbiome, TLR2, and the hygiene hypothesis. Autoimmun Rev 19(1):102430. https://doi.org/10.1016/j.autrev.2019.102430
Yuan D, Huang B, Gu M, Qin B-E, Su Z, Dai K, Peng F-H, Jiang Y (2023) Exploring shared genetic signatures of Alzheimer’s disease and multiple sclerosis: a bioinformatic analysis study. European Neurology
Kiefer L, Chiosso A, Langen J, Buckley A, Gaudin S, Rajkumar SM, Servito GIF, Cha ES, Vijay A, Yeung A, Horta A, Mui MH, Canzio D (2023) WAPL functions as a rheostat of Protocadherin isoform diversity that controls neural wiring. Science 380(6651):eadf8440. https://doi.org/10.1126/science.adf8440
Coleman JH, Lin B, Schwob JE (2017) Dissecting LSD1-dependent neuronal maturation in the olfactory epithelium. J Comp Neurol 525(16):3391–3413. https://doi.org/10.1002/cne.24259
Takaki E, Fujimoto M, Sugahara K, Nakahari T, Yonemura S, Tanaka Y, Hayashida N, Inouye S, Takemoto T, Yamashita H, Nakai A (2006) Maintenance of olfactory neurogenesis requires HSF1, a major heat shock transcription factor in mice. J Biol Chem 281(8):4931–4937. https://doi.org/10.1074/jbc.M506911200
Peng Q, Mechanic J, Shoieb A, Pardo ID, Schaevitz L, Fenyk-Melody J, Vitsky A, Boucher M, Somps C, Cook JC, Liu CN (2019) Circulating microRNA and automated motion analysis as novel methods of assessing chemotherapy-induced peripheral neuropathy in mice. PLoS One 14(1):e0210995. https://doi.org/10.1371/journal.pone.0210995
Paraboschi EM, Soldà G, Gemmati D, Orioli E, Zeri G, Benedetti MD, Salviati A, Barizzone N, Leone M, Duga S, Asselta R (2011) Genetic association and altered gene expression of mir-155 in multiple sclerosis patients. Int J Mol Sci 12(12):8695–8712. https://doi.org/10.3390/ijms12128695
Jevtić B, Timotijević G, Stanisavljević S, Momčilović M, Mostarica Stojković M, Miljković D (2015) Micro RNA-155 participates in re-activation of encephalitogenic T cells. Biomed Pharmacother 74:206–210. https://doi.org/10.1016/j.biopha.2015.08.011
Author Contributions
Conceptualization, T.S. and C.M.; methodology, J.K., K.J., and M.A.; investigation, T.S., S.H., and J.K.; writing—original draft, S.H.; writing—review and editing, C.M., C.A., and P.S.Q.; funding acquisition, T.S.
Funding
This research was supported by the 2024 scientific promotion program funded by Jeju National University. The funders had no role in the design, analysis, write-up, or decision to submit for publication.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics Approval
All experimental procedures were conducted in accordance with the Guidelines for the Care and Use of Laboratory Animals of Jeju National University (Nos. 2017-0019, 2022-0062). The animal protocols also conformed to the National Institutes of Health Guide for the Care and Use of Laboratory Animals (No. 85-23; 1985, rev. 1996).
Consent to Participate
Not applicable
Consent for Publication
Not applicable
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hong, S., Kim, J., Ahn, M. et al. Key Genes in Olfactory Disorder in Experimental Autoimmune Encephalomyelitis Identified by Transcriptomic Analysis of the Olfactory Bulbs. Mol Neurobiol (2024). https://doi.org/10.1007/s12035-024-03923-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12035-024-03923-0