Abstract
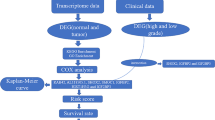
Recent researches reported that neurotrophins can promote glioma growth/invasion but the relevant model for predicting patients’ survival in Lower-Grade Gliomas (LGGs) lacked. In this study, we adopted univariate Cox analysis, LASSO regression, and multivariate Cox analysis to determine a signature including five neurotrophin-related genes (NTGs), CLIC1, SULF2, TGIF1, TTF2, and WEE1. Two-sample Mendelian Randomization (MR) further explored whether these prognostic-related genes were genetic variants that increase the risk of glioma. A total of 1306 patients have been included in this study, and the results obtained from the training set can be verified by four independent validation sets. The low-risk subgroup had longer overall survival in five datasets, and its AUC values all reached above 0.7. The risk groups divided by the NTGs signature exhibited a distinct difference in targeted therapies from the copy-number variation, somatic mutation, LGG’s surrounding microenvironment, and drug response. MR corroborated that TGIF1 was a potential causal target for increasing the risk of glioma. Our study identified a five-NTGs signature that presented an excellent survival prediction and potential biological function, providing new insight for the selection of LGGs therapy.









Similar content being viewed by others
Availability of Data and Materials
The datasets generated and analyzed during the current study are available in the Cancer Genome Atlas (TCGA) repository, (https://portal.gdc.cancer.gov/), the GSEA database (http://software.broadinstitute.org/gsea/msigdb/index.jsp), ArrayExpress database (https://www.ebi.ac.uk/arrayexpress/), and the Chinese Glioma Genome Atlas (CGGA) repository, (http://www.cgga.org.cn/).
Abbreviations
- LGGs:
-
Lower-Grade Gliomas
- NTGs:
-
Neurotrophin-related genes
- TME:
-
Tumor microenvironment
- NGF:
-
Nerve growth factor
- BDNF:
-
Brain-derived neurotrophic factor
- NT-3:
-
Neurotrophin-3
- p75NTR:
-
P75 neurotrophin receptor
- TCGA:
-
The Cancer Genome Atlas
- CGGA:
-
Chinese Glioma Genome Atlas
- TPM:
-
Transcript per million
- LASSO:
-
Least absolute shrinkage and selection operator
- KM:
-
Kaplan–Meier
- ssGSEA:
-
Single-sample gene set enrichment
- TMB:
-
Tumor mutational burden
- DEGs:
-
Differentially expressed genes
- ICIs:
-
Immune checkpoint inhibitors
- GEP:
-
Gene expression signature
- MIAS:
-
MHC I association immunoscore
- GDSC:
-
Genomics of Drug Sensitivity in Cancer
- GSA:
-
Gene set analysis
- PCA:
-
Principal component analysis
- IC50:
-
The half-maximal inhibitory concentration
References
Wen, P. Y., & Kesari, S. (2008). Malignant gliomas in adults. New England Journal of Medicine, 359(5), 492–507. https://doi.org/10.1056/NEJMra0708126
Fuller, G. N. (2008). The WHO Classification of Tumours of the Central Nervous System, 4th edition. Arch Pathology Laboratory Medicine, 132(6), 906. https://doi.org/10.5858/2008-132-906-TWCOTO
Ostrom, Q. T., Gittleman, H., Farah, P., et al. (2013). CBTRUS statistical report: Primary brain and central nervous system tumors diagnosed in the United States in 2006–2010. Neuro-Oncology., 15(2), 1–56. https://doi.org/10.1093/neuonc/not151
Cancer Genome Atlas Research Network, Brat, D. J., Verhaak, R. G. W., et al. (2015). Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. New England Journal of Medicine, 372(26), 2481–2498. https://doi.org/10.1056/NEJMoa1402121
Lactobacillus rhamnosus GG induces cGAS/STING- dependent type I interferon and improves response to immune checkpoint blockade - PubMed. Retrieved January 30, 2023, from https://pubmed.ncbi.nlm.nih.gov/33685966/
S100A gene family: immune-related prognostic biomarkers and therapeutic targets for low-grade glioma - PubMed. Retrieved January 30, 2023, from https://pubmed.ncbi.nlm.nih.gov/34148033/
Hypoxia-Related lncRNA Correlates With Prognosis and Immune Microenvironment in Lower-Grade Glioma - PubMed. Retrieved January 30, 2023, from https://pubmed.ncbi.nlm.nih.gov/34659218/
Tao, B., Ling, Y., Zhang, Y., et al. (2019). CA10 and CA11 negatively regulate neuronal activity-dependent growth of gliomas. Molecular Oncology, 13(5), 1018–1032. https://doi.org/10.1002/1878-0261.12445
Hamel, W., Westphal, M., Szönyi, E., Escandón, E., & Nikolics, K. (1993). Neurotrophin gene expression by cell lines derived from human gliomas. Journal of Neuroscience Research, 34(2), 147–157. https://doi.org/10.1002/jnr.490340202
Venkatesh, H. S., Johung, T. B., Caretti, V., et al. (2015). Neuronal activity promotes glioma growth through neuroligin-3 secretion. Cell, 161(4), 803–816. https://doi.org/10.1016/j.cell.2015.04.012
Venkatesh, H. S., Tam, L. T., Woo, P. J., et al. (2017). Targeting neuronal activity-regulated neuroligin-3 dependency in high-grade glioma. Nature, 549(7673), 533–537. https://doi.org/10.1038/nature24014
Calatozzolo, C., Salmaggi, A., Pollo, B., et al. (2007). Expression of cannabinoid receptors and neurotrophins in human gliomas. Neurological Sciences: Official Journal of the Italian Neurological Society and of the Italian Society of Clinical Neurophysiology, 28(6), 304–310. https://doi.org/10.1007/s10072-007-0843-8
Wang, X., Prager, B. C., Wu, Q., et al. (2018). Reciprocal signaling between glioblastoma stem cells and differentiated tumor cells promotes malignant progression. Cell Stem Cell, 22(4), 514-528.e5. https://doi.org/10.1016/j.stem.2018.03.011
Johnston, A. L. M., Lun, X., Rahn, J. J., et al. (2007). The p75 neurotrophin receptor is a central regulator of glioma invasion. PLoS Biology, 5(8), e212. https://doi.org/10.1371/journal.pbio.0050212
Johung, T., & Monje, M. (2017). Neuronal activity in the glioma microenvironment. Current Opinion in Neurobiology, 47, 156–161. https://doi.org/10.1016/j.conb.2017.10.009
Lai, G., Zhong, X., Liu, H., Deng, J., Li, K., & Xie, B. (2022). Development of a hallmark pathway-related gene signature associated with immune response for lower grade gliomas. International Journal of Molecular Sciences, 23(19), 11971. https://doi.org/10.3390/ijms231911971
Charoentong, P., Finotello, F., Angelova, M., et al. (2017). Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Reports, 18(1), 248–262. https://doi.org/10.1016/j.celrep.2016.12.019
Auslander, N., Zhang, G., Lee, J. S., et al. (2018). Robust prediction of response to immune checkpoint blockade therapy in metastatic melanoma. Nature Medicine, 24(10), 1545–1549. https://doi.org/10.1038/s41591-018-0157-9
Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy - PubMed. Retrieved January 30, 2023, from https://pubmed.ncbi.nlm.nih.gov/30309915/
Wu, C. C., Wang, Y. A., Livingston, J. A., Zhang, J., & Futreal, P. A. (2022). Prediction of biomarkers and therapeutic combinations for anti-PD-1 immunotherapy using the global gene network association. Nature Communications, 13(1), 42. https://doi.org/10.1038/s41467-021-27651-4
Yang, W., Soares, J., Greninger, P., et al. (2013). Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Research, 41(1), 955–961. https://doi.org/10.1093/nar/gks1111
Liu, C. J., Hu, F. F., Xie, G. Y., et al. (2023). GSCA: an integrated platform for gene set cancer analysis at genomic, pharmacogenomic and immunogenomic levels. Brief Bioinformatics, 24(1), bbac558. https://doi.org/10.1093/bib/bbac558
Mermel, C. H., Schumacher, S. E., Hill, B., Meyerson, M. L., Beroukhim, R., & Getz, G. (2011). GISTIC20 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biology, 12(4), 41. https://doi.org/10.1186/gb-2011-12-4-r41
Birnbaum, T., Roider, J., Schankin, C. J., et al. (2007). Malignant gliomas actively recruit bone marrow stromal cells by secreting angiogenic cytokines. Journal of Neuro-oncology, 83(3), 241–247. https://doi.org/10.1007/s11060-007-9332-4
Li, R. K., Zhang, J., Zhang, Y. H., Li, M. L., Wang, M., & Tang, J. W. (2012). Chloride intracellular channel 1 is an important factor in the lymphatic metastasis of hepatocarcinoma. Biomedicine & Pharmacotherapy, 66(3), 167–172. https://doi.org/10.1016/j.biopha.2011.10.002
Petrova, D. T., Asif, A. R., Armstrong, V. W., et al. (2008). Expression of chloride intracellular channel protein 1 (CLIC1) and tumor protein D52 (TPD52) as potential biomarkers for colorectal cancer. Clinical Biochemistry, 41(14–15), 1224–1236. https://doi.org/10.1016/j.clinbiochem.2008.07.012
Wang, J. W., Peng, S. Y., Li, J. T., et al. (2009). Identification of metastasis-associated proteins involved in gallbladder carcinoma metastasis by proteomic analysis and functional exploration of chloride intracellular channel 1. Cancer Letters, 281(1), 71–81. https://doi.org/10.1016/j.canlet.2009.02.020
Zhang, S., Wang, X. M., Yin, Z. Y., et al. (2013). Chloride intracellular channel 1 is overexpression in hepatic tumor and correlates with a poor prognosis. APMIS Acta Pathology Microbiology Immunology Scandinavia, 121(11), 1047–1053. https://doi.org/10.1111/apm.12093
Chang, Y. H., Wu, C. C., Chang, K. P., Yu, J. S., Chang, Y. C., & Liao, P. C. (2009). Cell secretome analysis using hollow fiber culture system leads to the discovery of CLIC1 protein as a novel plasma marker for nasopharyngeal carcinoma. Journal of Proteome Research, 8(12), 5465–5474. https://doi.org/10.1021/pr900454e
Chen, C. D., Wang, C. S., Huang, Y. H., et al. (2007). Overexpression of CLIC1 in human gastric carcinoma and its clinicopathological significance. Proteomics, 7(1), 155–167. https://doi.org/10.1002/pmic.200600663
Peng, J. M., Lin, S. H., Yu, M. C., & Hsieh, S. Y. (2021). CLIC1 recruits PIP5K1A/C to induce cell-matrix adhesions for tumor metastasis. Journal of Clinical Investigation., 131(1), e133525. https://doi.org/10.1172/JCI133525
Wang, L., He, S., Tu, Y., et al. (2012). Elevated expression of chloride intracellular channel 1 is correlated with poor prognosis in human gliomas. Journal of Experimental Clinical Cancer Research Centre, 31(1), 44. https://doi.org/10.1186/1756-9966-31-44
Phillips, J. J., Huillard, E., Robinson, A. E., et al. (2012). Heparan sulfate sulfatase SULF2 regulates PDGFRα signaling and growth in human and mouse malignant glioma. The Journal of Clinical Investigation, 122(3), 911–922. https://doi.org/10.1172/JCI58215
Wang, B., Ma, Q., Wang, X., Guo, K., Liu, Z., & Li, G. (2022). TGIF1 overexpression promotes glioma progression and worsens patient prognosis. Cancer Medicine, 11(24), 5113–5128. https://doi.org/10.1002/cam4.4822
Wu, S., Wang, S., Gao, F., et al. (2018). Activation of WEE1 confers resistance to PI3K inhibition in glioblastoma. Neuro-Oncology, 20(1), 78–91. https://doi.org/10.1093/neuonc/nox128
Hanahan, D., & Weinberg, R. A. (2011). Hallmarks of cancer: The next generation. Cell, 144(5), 646–674. https://doi.org/10.1016/j.cell.2011.02.013
Dubois, F., Sidiropoulos, N., Weischenfeldt, J., & Beroukhim, R. (2022). Structural variations in cancer and the 3D genome. Nature Reviews Cancer, 22(9), 533–546. https://doi.org/10.1038/s41568-022-00488-9
Chae, M., Peterson, T. E., Balgeman, A., et al. (2015). Increasing glioma-associated monocytes leads to increased intratumoral and systemic myeloid-derived suppressor cells in a murine model. Neuro-Oncology, 17(7), 978–991. https://doi.org/10.1093/neuonc/nou343
Zhang, N., Dai, Z., Wu, W., et al. (2021). The predictive value of monocytes in immune microenvironment and prognosis of glioma patients based on machine learning. Frontiers in Immunology, 12, 656541. https://doi.org/10.3389/fimmu.2021.656541
Chan, T. A., Wolchok, J. D., & Snyder, A. (2015). Genetic basis for clinical response to CTLA-4 blockade in melanoma. New England Journal of Medicine, 373(20), 1984. https://doi.org/10.1056/NEJMc1508163
Rizvi, N. A., Hellmann, M. D., Snyder, A., et al. (2015). Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science, 348(6230), 124–128. https://doi.org/10.1126/science.aaa1348
Hugo, W., Zaretsky, J. M., Sun, L., et al. (2017). Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell, 168(3), 542. https://doi.org/10.1016/j.cell.2017.01.010
Carbone, D. P., Reck, M., Paz-Ares, L., et al. (2017). First-Line Nivolumab in Stage IV or Recurrent Non-Small-Cell Lung Cancer. New England Journal of Medicine, 376(25), 2415–2426. https://doi.org/10.1056/NEJMoa1613493
Choi, S., Yu, Y., Grimmer, M. R., Wahl, M., Chang, S. M., & Costello, J. F. (2018). Temozolomide-associated hypermutation in gliomas. Neuro-Oncology, 20(10), 1300–1309. https://doi.org/10.1093/neuonc/noy016
Yu, Y., Villanueva-Meyer, J., Grimmer, M. R., et al. (2021). Temozolomide-induced hypermutation is associated with distant recurrence and reduced survival after high-grade transformation of low-grade IDH-mutant gliomas. Neuro-Oncology, 23(11), 1872–1884. https://doi.org/10.1093/neuonc/noab081
Fangusaro, J., Onar-Thomas, A., Young Poussaint, T., et al. (2019). Selumetinib in paediatric patients with BRAF-aberrant or neurofibromatosis type 1-associated recurrent, refractory, or progressive low-grade glioma: A multicentre, phase 2 trial. The Lancet Oncology, 20(7), 1011–1022. https://doi.org/10.1016/S1470-2045(19)30277-3
Perreault, S., Larouche, V., Tabori, U., et al. (2019). A phase 2 study of trametinib for patients with pediatric glioma or plexiform neurofibroma with refractory tumor and activation of the MAPK/ERK pathway: TRAM-01. BMC Cancer, 19(1), 1250. https://doi.org/10.1186/s12885-019-6442-2
Jane, E. P., Premkumar, D. R., Rajasundaram, D., et al. (2022). Reversing tozasertib resistance in glioma through inhibition of pyruvate dehydrogenase kinases. Molecular Oncology, 16(1), 219–249. https://doi.org/10.1002/1878-0261.13025
Yang, Y., Lei, T., Du, S., et al. (2018). Nuclear GSK3β induces DNA double-strand break repair by phosphorylating 53BP1 in glioblastoma. International Journal of Oncology, 52(3), 709–720. https://doi.org/10.3892/ijo.2018.4237
Acknowledgements
We are very grateful to the TCGA, CGGA, GSEA, and CIBERSOR database and other public resources for providing us with a research foundation.
Funding
This research was funded by (National Youth Science Foundation Project), grant number [82204159]) and Science and Technology Research Program of Chongqing Municipal Education Commission (Grant No.KJQN202300423).
Author information
Authors and Affiliations
Contributions
Conceptualization, CZ; methodology, CZ and JD; software, CZ and KL; validation, CZ and JD; formal analysis, CZ and LC; resources, GL; data curation, GL; writing—original draft preparation, CZ; writing—review and editing, XZ and BX; visualization, CZ; supervision, BX; project administration, XZ; funding acquisition, BX. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Ethical approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, C., Lai, G., Deng, J. et al. Integrating Machine Learning and Mendelian Randomization Determined a Functional Neurotrophin-Related Gene Signature in Patients with Lower-Grade Glioma. Mol Biotechnol (2024). https://doi.org/10.1007/s12033-023-01045-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12033-023-01045-x