Abstract
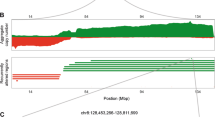
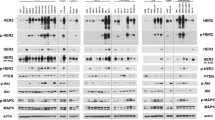
Breast cancer is one of the leading causes of cancer death in women, and although treatment outcome has substantially improved in the past decades, advanced or metastatic breast cancers still carry a poor prognosis. Gene amplification is one of the frequent genetic alterations in cancer, and oncogene amplification may be associated with cancer aggressiveness and oncogenicity. Targeting amplified genes such as HER2 has vastly improved disease outcome and survival, and anti-HER2 therapeutics have revolutionized the standard of care in HER2 breast cancer. Besides currently known druggable gene amplifications including ERBB2 and FGFR2, other frequently amplified genes are relatively less well known for function and clinical significance. By querying four large databases from TCGA and AACR-Genie, from a total of 11,890 patients with invasive ductal breast carcinoma, we discover IKZF3, CCND1, ERBB2 to be consistently amplified across different cohorts. We further identify IKZF3 as a frequently amplified gene in breast cancer with a prevalence of 12–15% amplification rate. Interestingly, IKZF3 amplification is frequently co-amplified with ERBB2/HER2, and is also associated with worse prognosis compared to IKZF3 non-amplified cancers. Analysis of HER2 breast cancer patients treated with trastuzumab revealed decrease in both ERBB2/HER2 and IKZF3 expression. Further investigation using the DepMap for gene dependency by genome-wide CRISPR screening revealed dependence on IKZF3 in HER2 breast cancer cell lines. Our study utilized an integrative analysis of large-scale patient genomics, transcriptomics and clinical data to reveal IKZF3 as a frequently amplified gene, and suggest a potential role of IKZF3 as a druggable target for HER2 breast cancer.




Similar content being viewed by others
Abbreviations
- BC:
-
Breast cancer
- DAVID:
-
Database for Annotation, Visualization, and Integrated Discovery
- DFS:
-
Disease-free survival
- GEPIA:
-
Gene Expression Profiling Interactive Analysis
- GO:
-
Gene ontology
- HER2:
-
Epidermal growth factor receptor 2
- IMiDs:
-
Immunomodulatory imide drugs
- IPA:
-
Ingenuity pathway analysis
- IDC:
-
Invasive ductal breast carcinoma
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- mBC:
-
Metastatic breast cancer
- OS:
-
Overall survival
- PFS:
-
Progression-free survival
- PROTACs:
-
Proteolysis targeting chimeras
- TCGA:
-
The Cancer Genome Atlas
References
De Cicco P, Catani MV, Gasperi V, Sibilano M, Quaglietta M, Savini I. Nutrition and breast cancer: a literature review on prevention. Treat Recurr Nutr. 2019;11(7):1514. https://doi.org/10.3390/nu11071514.
Ademuyiwa FO, Edge SB, Erwin DO, Orom H, Ambrosone CB, Underwood W 3rd. Breast cancer racial disparities: unanswered questions. Cancer Res. 2011;71(3):640–4. https://doi.org/10.1158/0008-5472.CAN-10-3021.
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. https://doi.org/10.3322/caac.21442.
Swain SM, Miles D, Kim SB, Im YH, Im SA, Semiglazov V, et al. Pertuzumab, trastuzumab, and docetaxel for HER2-positive metastatic breast cancer (CLEOPATRA): end-of-study results from a double-blind, randomised, placebo-controlled, phase 3 study. Lancet Oncol. 2020;21(4):519–30. https://doi.org/10.1016/S1470-2045(19)30863-0.
Hortobagyi GN, Stemmer SM, Burris HA, Yap YS, Sonke GS, Hart L, Campone M, Petrakova K, Winer EP, Janni W, Conte,. LBA17 Overall survival (OS) results from the phase III MONALEESA-2 (ML-2) trial of postmenopausal patients (pts) with hormone receptor positive/human epidermal growth factor receptor 2 negative (HR+/HER2−) advanced breast cancer (ABC) treated with endocrine therapy (ET) ± ribociclib (RIB). Ann Oncol. 2021. https://doi.org/10.1016/j.annonc.2021.08.2090.
Albertson DG. Gene amplification in cancer. Trends Genet. 2006;22(8):447–55. https://doi.org/10.1016/j.tig.2006.06.007.
Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70.
Albertson DG, Collins C, McCormick F, Gray JW. Chromosome aberrations in solid tumors. Nat Genet. 2003;34(4):369–76. https://doi.org/10.1038/ng1215.
Al-Kuraya K, Schraml P, Torhorst J, Tapia C, Zaharieva B, Novotny H, et al. Prognostic relevance of gene amplifications and coamplifications in breast cancer. Can Res. 2004;64(23):8534–40. https://doi.org/10.1158/0008-5472.CAN-04-1945.
Kato S, Okamura R, Mareboina M, Lee S, Goodman A, Patel SP, et al. Revisiting epidermal growth factor receptor (EGFR) amplification as a target for anti-EGFR therapy: analysis of cell-free circulating tumor DNA in patients with advanced malignancies. JCO Precis Oncol. 2019. https://doi.org/10.1200/PO.18.00180.
Saborowski A, Lehmann U, Vogel A. FGFR inhibitors in cholangiocarcinoma: what’s now and what’s next? Ther Adv Med Oncol. 2020;12:1758835920953293. https://doi.org/10.1177/1758835920953293.
Heizmann B, Kastner P, Chan S. The Ikaros family in lymphocyte development. Curr Opin Immunol. 2018;51:14–23. https://doi.org/10.1016/j.coi.2017.11.005.
Sievers QL, Petzold G, Bunker RD, Renneville A, Slabicki M, Liddicoat BJ, et al. Defining the human C2H2 zinc finger degrome targeted by thalidomide analogs through CRBN. Science. 2018. https://doi.org/10.1126/science.aat0572.
Chen K, Quan J, Yang J, Chen Z. The potential markers of endocrine resistance among HR+ /HER2+ breast cancer patients. Clin transl oncol. 2020;22(4):576–84. https://doi.org/10.1007/s12094-019-02163-2.
Sircoulomb F, Bekhouche I, Finetti P, Adelaide J, Ben Hamida A, Bonansea J, et al. Genome profiling of ERBB2-amplified breast cancers. BMC Cancer. 2010;10:539. https://doi.org/10.1186/1471-2407-10-539.
Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6(1):1–6. https://doi.org/10.1016/s1476-5586(04)80047-2.
Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–102. https://doi.org/10.1093/nar/gkx247.
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–4. https://doi.org/10.1158/2159-8290.CD-12-0095.
da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. https://doi.org/10.1038/nprot.2008.211.
Huang DW, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucl acid res. 2007. https://doi.org/10.1093/nar/gkm415.
Atlas CG, N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. https://doi.org/10.1038/nature11412.
Consortium APG. AACR project GENIE: powering precision medicine through an international consortium. Cancer Discov. 2017;7(8):818–31. https://doi.org/10.1158/2159-8290.CD-17-0151.
Frampton GM, Fichtenholtz A, Otto GA, Wang K, Downing SR, He J, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31(11):1023–31. https://doi.org/10.1038/nbt.2696.
Consortium APG. Metastatic Breast Cancer: 2013–2016 (DFCI, CCR 2020).
Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–52. https://doi.org/10.1038/nature10983.
Pereira B, Chin SF, Rueda OM, Vollan HK, Provenzano E, Bardwell HA, et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun. 2016;7:11479. https://doi.org/10.1038/ncomms11479.
AACR-Genie. AACR GENIE Data Guide.
Lundberg A, Lindstrom LS, Li J, Harrell JC, Darai-Ramqvist E, Sifakis EG, et al. The long-term prognostic and predictive capacity of cyclin D1 gene amplification in 2305 breast tumours. Breast Cancer Res. 2019;21(1):34. https://doi.org/10.1186/s13058-019-1121-4.
Finn RS, Crown JP, Lang I, Boer K, Bondarenko IM, Kulyk SO, et al. The cyclin-dependent kinase 4/6 inhibitor palbociclib in combination with letrozole versus letrozole alone as first-line treatment of oestrogen receptor-positive, HER2-negative, advanced breast cancer (PALOMA-1/TRIO-18): a randomised phase 2 study. Lancet Oncol. 2015;16(1):25–35. https://doi.org/10.1016/S1470-2045(14)71159-3.
Modi S, Saura C, Yamashita T, Park YH, Kim SB, Tamura K, et al. Trastuzumab deruxtecan in previously treated HER2-positive breast cancer. N Engl J Med. 2020;382(7):610–21. https://doi.org/10.1056/NEJMoa1914510.
Swain SM, Baselga J, Kim SB, Ro J, Semiglazov V, Campone M, et al. Pertuzumab, trastuzumab, and docetaxel in HER2-positive metastatic breast cancer. N Engl J Med. 2015;372(8):724–34. https://doi.org/10.1056/NEJMoa1413513.
Bivin WW, Yergiyev O, Bunker ML, Silverman JF, Krishnamurti U. GRB7 expression and correlation With HER2 amplification in invasive breast carcinoma. Appl Immunohistochem Mol Morphol. 2017;25(8):553–8. https://doi.org/10.1097/PAI.0000000000000349.
Triulzi T, Regondi V, De Cecco L, Cappelletti MR, Di Modica M, Paolini B, et al. Early immune modulation by single-agent trastuzumab as a marker of trastuzumab benefit. Br J Cancer. 2018;119(12):1487–94. https://doi.org/10.1038/s41416-018-0318-0.
Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS, et al. Defining a cancer dependency map. Cell. 2017;170(3):564–76. https://doi.org/10.1016/j.cell.2017.06.010.
Perez EA, Romond EH, Suman VJ, Jeong JH, Sledge G, Geyer CE Jr, et al. Trastuzumab plus adjuvant chemotherapy for human epidermal growth factor receptor 2-positive breast cancer: planned joint analysis of overall survival from NSABP B-31 and NCCTG N9831. J Clin Oncol Off J American Soc Clin Oncol. 2014;32(33):3744–52. https://doi.org/10.1200/JCO.2014.55.5730.
Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344(11):783–92. https://doi.org/10.1056/NEJM200103153441101.
Cortés J, Kim SB, Chung WP, Im SA, Park YH, Hegg R, Kim MH, Tseng LM, Petry V, Chung CF, Iwata H. LBA1 - Trastuzumab deruxtecan (T-DXd) vs trastuzumab emtansine (T-DM1) in patients (Pts) with HER2+ metastatic breast cancer (mBC): Results of the randomized phase III DESTINY-Breast03 study. Ann Oncol. 2021. https://doi.org/10.1016/j.annonc.2021.08.2087.
Shao Z, Tseng LM, Huang CS, Pang D, Yang Y, Li W, et al. Pertuzumab and trastuzumab as adjuvant treatment for HER2-positive early breast cancer: outcomes in Chinese patients in the APHINITY study. Jpn J Clin Oncol. 2021;51(3):345–53. https://doi.org/10.1093/jjco/hyaa216.
Piccart M, Procter M, Fumagalli D, de Azambuja E, Clark E, Ewer MS, et al. Adjuvant pertuzumab and trastuzumab in early HER2-positive breast cancer in the APHINITY Trial: 6 years’ follow-up. J Clin Oncol. 2021;39(13):1448–57. https://doi.org/10.1200/JCO.20.01204.
Menard S, Pupa SM, Campiglio M, Tagliabue E. Biologic and therapeutic role of HER2 in cancer. Oncogene. 2003;22(42):6570–8. https://doi.org/10.1038/sj.onc.1206779.
Patel A, Unni N, Peng Y. The changing paradigm for the treatment of HER2-positive breast cancer. Cancers (Basel). 2020. https://doi.org/10.3390/cancers12082081.
Ferrari A, Vincent-Salomon A, Pivot X, Sertier AS, Thomas E, Tonon L, et al. A whole-genome sequence and transcriptome perspective on HER2-positive breast cancers. Nat Commun. 2016;7:12222. https://doi.org/10.1038/ncomms12222.
Kronke J, Udeshi ND, Narla A, Grauman P, Hurst SN, McConkey M, et al. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science. 2014;343(6168):301–5. https://doi.org/10.1126/science.1244851.
Lu G, Middleton RE, Sun H, Naniong M, Ott CJ, Mitsiades CS, et al. The myeloma drug lenalidomide promotes the cereblon-dependent destruction of Ikaros proteins. Science. 2014;343(6168):305–9. https://doi.org/10.1126/science.1244917.
Furihata H, Yamanaka S, Honda T, Miyauchi Y, Asano A, Shibata N, et al. Structural bases of IMiD selectivity that emerges by 5-hydroxythalidomide. Nat Commun. 2020;11(1):4578. https://doi.org/10.1038/s41467-020-18488-4.
Steinebach C, Lindner S, Udeshi ND, Mani DC, Kehm H, Kopff S, et al. Homo-PROTACs for the chemical knockdown of cereblon. ACS Chem Biol. 2018;13(9):2771–82. https://doi.org/10.1021/acschembio.8b00693.
Sun X, Gao H, Yang Y, He M, Wu Y, Song Y, et al. PROTACs: great opportunities for academia and industry. Signal Transduct Target Ther. 2019;4:64. https://doi.org/10.1038/s41392-019-0101-6.
Barrio S, Munawar U, Zhu YX, Giesen N, Shi CX, Via MD, et al. IKZF1/3 and CRL4(CRBN) E3 ubiquitin ligase mutations and resistance to immunomodulatory drugs in multiple myeloma. Haematologica. 2020;105(5):e237–41. https://doi.org/10.3324/haematol.2019.217943.
Khan S, He Y, Zhang X, Yuan Y, Pu S, Kong Q, et al. PROteolysis TArgeting Chimeras (PROTACs) as emerging anticancer therapeutics. Oncogene. 2020;39(26):4909–24. https://doi.org/10.1038/s41388-020-1336-y.
Yin LL, Wen XM, Lai QH, Li J, Wang XW. Lenalidomide improvement of cisplatin antitumor efficacy on triple-negative breast cancer cells in vitro. Oncol Lett. 2018;15(5):6469–74. https://doi.org/10.3892/ol.2018.8120.
Brosseau C, Colston K, Dalgleish AG, Galustian C. The immunomodulatory drug lenalidomide restores a vitamin D sensitive phenotype to the vitamin D resistant breast cancer cell line MDA-MB-231 through inhibition of BCL-2: potential for breast cancer therapeutics. Apoptosis : Int J Program cell death. 2012;17(2):164–73. https://doi.org/10.1007/s10495-011-0670-5.
Barbarossa A, Iacopetta D, Sinicropi MS, Franchini C, Carocci A. Recent advances in the development of thalidomide-related compounds as anticancer drugs. Curr Med Chem. 2021. https://doi.org/10.2174/0929867328666210623143526.
Wang X, Shen Y, MengLv L, Zhang X, Yang J, Wang F, et al. Thalidomide suppresses breast cancer tumor growth by inhibiting tumor-associated macrophage accumulation in breast tumor-bearing mice. Eur J Pharm Sci. 2020;151: 105302. https://doi.org/10.1016/j.ejps.2020.105302.
Biran A, Yin S, Kretzmer H, Ten Hacken E, Parvin S, Lucas F, et al. Activation of notch and Myc signaling via B cell-restricted depletion of Dnmt3a generates a consistent murine model of chronic lymphocytic leukemia. Can Res. 2021. https://doi.org/10.1158/0008-5472.CAN-21-1273.
Lazarian G, Yin S, Ten Hacken E, Sewastianik T, Uduman M, Font-Tello A, et al. A hotspot mutation in transcription factor IKZF3 drives B cell neoplasia via transcriptional dysregulation. Cancer Cell. 2021;39(3):380–93. https://doi.org/10.1016/j.ccell.2021.02.003.
Razavi P, Chang MT, Xu G, Bandlamudi C, Ross DS, Vasan N, et al. The genomic landscape of endocrine-resistant advanced breast cancers. Cancer Cell. 2018;34(3):427–38. https://doi.org/10.1016/j.ccell.2018.08.008.
Razavi P, Dickler MN, Shah PD, Toy W, Brown DN, Won HH, et al. Alterations in PTEN and ESR1 promote clinical resistance to alpelisib plus aromatase inhibitors. Nat Cancer. 2020;1(4):382–93. https://doi.org/10.1038/s43018-020-0047-1.
Li Q, Jiang B, Guo J, Shao H, Del Priore IS, Chang Q, et al. INK4 tumor suppressor proteins mediate resistance to CDK4/6 kinase inhibitors. Cancer Discov. 2021. https://doi.org/10.1158/2159-8290.CD-20-1726.
Acknowledgements
This study was partially funded by the grants to JIL: MOST 110-2314-B-A49A-549 (Ministry of Science and Technology), CI-109-8 (Yen Tjing Ling Medical Foundation), Taiwan Clinical Oncology Research Foundation, and 109DHA0100490 (Taipei Veterans General Hospital internal grant).
Funding
Hsinchu Science Park Bureau,Ministry of Science and Technology,Taiwan,MOST 110-2314-B-A49A-549,Jiun-I Lai
Author information
Authors and Affiliations
Contributions
CJY, CIS, CYL, JIL: data collection, analysis and curation. CJY, CYL, JIL: drafting of the manuscript. CYL, TCC, CCH, LMT, JIL: conceptualization of the project. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lin, CY., Yu, CJ., Shen, CI. et al. IKZF3 amplification frequently occurs in HER2-positive breast cancer and is a potential therapeutic target. Med Oncol 39, 242 (2022). https://doi.org/10.1007/s12032-022-01812-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-022-01812-x