Abstract
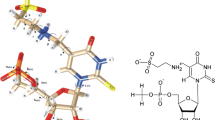
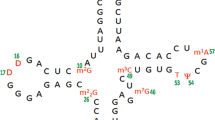
Structural significance of conformational preferences and ribose ring puckering of newly discovered hyper modified nucleotide, 5’-monophosphate 2-methylthio cyclic N6-threonylcarbamoyladenosine (p-ms2ct6A) have been investigated using quantum chemical semi-empirical RM1 and molecular dynamics simulation techniques. Automated geometry optimization of most stable structure of p-ms2ct6A has also been carried out with the help of abinitio (HF SCF, DFT) as well as semi empirical quantum chemical (RM1, AM1, PM3, and PM6) methods. Most stable structure of p-ms2ct6A is stabilized by intramolecular interactions between N(3)…HC(2’), N(1)…HC(16), O(13)…HC(15), and O(13)…HO(14). The torsion angles alpha (α) and beta (β) show the significant characteristic patterns with the involvement of intramolecular hydrogen bonding to provide stability to the p-ms2ct6A. Further, molecular dynamics simulations of p-ms2ct6A revealed the role of ribose sugar ring puckering i.e. C2’-endo and C3’-endo on the structural dynamics of ms2ct6A side chain. The modified nucleotide p-ms2ct6A periodically prefers both the C2’-endo and C3’-endo sugar with ‘anti’ and ‘syn’ conformations. This property of p-ms2ct6A could be useful to recognize the starting ANN codons. All atom explicit MD simulation of anticodon loop (ACL) of tRNALys of Bacillus subtilis containing ms2ct6A at 37th position showed the U-turn feature, base stacking ability with other adjacent bases and hydrogen bonding interactions similar to the isolated base p-ms2ct6A. The ribose sugar puckering contributes to the orientation of the side chain conformation of p-ms2ct6A. Thus, the present study could be helpful to understand the structure-function relationship of the hypermodified nucleoside, ms2ct6A in recognition of the proper codons AAA/AAG during protein biosynthesis.








Similar content being viewed by others
References
Murphy, IV, F. V., Ramakrishnan, V., Malkiewicz, A., & Agris, P. F. (2004). The role of modifications in codon discrimination by tRNA LysUUU. Nature Structural & Molecular Biology, 11(12), 1186–1191.
Stuart, J. W., Gdaniec, Z., Guenther, R., Marszalek, M., Sochacka, E., Malkiewicz, A., & Agris, P. F. (2000). Functional anticodon architecture of human tRNALys3 includes disruption of intraloop hydrogen bonding by the naturally occurring amino acid modification, t6A. Biochemistry, 39, 13396–13404.
Moazed, D., & Noller, H. F. (1986). Transfer RNA shields specific nucleotides in 16S ribosomal RNA from attack by chemical probes. Cell, 47, 985–994.
Nguyen, H. A., Hoffer, E. D., & Dunham, C. M. (2019). Importance of a tRNA anticodon loop modification and a conserved, noncanonical anticodon stem pairing in tRNACGGPro for decoding. Journal of Biological Chemistry, 294(14), 5281–5291.
Jackman, J. E., & Alfonzo, J. D. (2013). Transfer RNA modification: nature’s combinatorial chemistry playground. Wiley Interdisciplinary Review. RNA, 4(1), 35–48.
Machnicka, M. A., Olchowik, A., Grosjean, H., & Bujnicki, J. M. (2014). Distribution and frequencies of post transcriptional modifications in tRNAs. RNA Biology, 11(12), 1619–1629.
Agris, P. F., Eruysal, E. R., Narendran, A., Vare, V. Y. P., & Vangaveti, S. (2018). Celebrating wobble decoding: Half a century and still much is new. RNA Biology, 15, 537–553.
Lin, C. A., Ellis, S. R., & True, H. L. (2010). The Sau5 protein is essential for normal translational regulation in yeast. Molecular and Cellular Biology, 30(1), 354–363.
Kang, B., Miyauchi, K., Matuszewski, M., D’Almeida, G. S., Rubio, M. A. T., Alfonzo, J. D., Inoue, K., Sakaguchi, K., Suzuki, T., Sochacka, E., & Suzuki, T. (2017). Identification of 2-methylthio cyclic N6-threonylcarbamoyladenosine (ms2ct6A) as a novel RNA modification at position 37 of tRNAs. Nucleic Acids Research, 45(4), 2124–2136.
Kamble, A. S., Sambhare, S. B., Fandilolu, P. M., & Sonwane, K. D. (2016). Structural significance of modified nucleoside 5-taurinomethyl-2- thiouridine, sm5s2U, found at ‘wobble’ position in anticodon loop of human mitochondrial tRNALys. Structural Chemistry, 27, 839–854.
Kumbhar, B. V., Kumbhar, N. M., & Sonawane, K. D. (2012). Conformational preferences and MD simulation studies of the hypermodified nucleic acid base ms2hn6Ade present at the 3’-adjacent (37th) position in the Anticodon loop of hyperthermophilic tRNAs. Internet Electronic Journal of Molecular Design, 11, 33–48.
Kumbhar, N. M., Kumbhar, B. V., & Sonawane, K. D. (2012). Structural significance of hypermodified nucleic acid base hydroxywybutine (OHyW) which occur at 37th position in the anticodon loop of yeast tRNAPhe. Journal of Molecular Graphics and Modelling, 38, 174–185.
Kamble A. S., Fandilolu P. M., Sambhare S. B., & Sonawane K. D. (2017). Idiosyncratic recognition of UUG/UUA codons by modified nucleoside 5-taurinomethyluridine, τm5U present at ‘wobble’ position in anticodon loop of tRNALeu: A molecular modeling approach. PLoS ONE. 1–16.
Fandilolu, P. M., Kamble, A. S., Sambhare, S. B., & Sonawane, K. D. (2018). Conformational preferences and structural analysis of hypermodified nucleoside, peroxywybutosine (o2yW) found at 37th position in anticodon loop of tRNAPhe and its role in modulating UUC codon-anticodon interactions. Gene, 641, 310–325.
Sambhare, S. B., Kumbhar, B. V., Kamble, A. D., Bavi, R. S., Kumbhar, N. M., & Sonawane, K. D. (2014). Structural significance of modified nucleosides k2Cand t6A present in the anticodon loop of tRNAIle. RSC Advances, 4, 14176–14188.
Yarus, M. (1982). Translational efficiency of Transfer RNA’s: Uses of an extended anticodon. Science, 218, 646–652.
Devi, M., & Duncan Lyngdoh, R. H. (2018). Favored and less favored codon-anticodon duplexes arising from the GC codon family box encoding for alanine: some computational perspectives. Journal of Biomolecular Structure and Dynamics, 36(4), 1029–1049.
Fuller, W., & Hodgson, A. (1967). Conformation of the Anticodon loop in tRNA. Nature, 215, 817–821.
Benas, P., Bec, G., Kieth, G., Marquet, R., Ehresmann, C., Ehresmann, B., & Dumas, P. (2000). The crystal structure of HIV reverse –transcription primer tRNA (Lys, 3) shows a canonical anticodon loop. RNA, 6, 1347–1355.
Sundaram, M., Crain, P. F., & Davis, D. R. (2000). Synthesis and characterization of the native anticodon domain of E.coli tRNALys: simultaneous incorporation of modified nucleosides mnm5s2U, t6A and Pseudouridine using phosphoramidite Chemistry. Journal of Organic Chemistry, 65, 5609–5614.
Miyauchi, K., Kimura, S., & Suzuki, T. (2012). A cyclic form of N6-threonylcarbamoyladenosine as a widely distributed tRNA hypermodification. Nature Chemical Biology, 9, 105–111.
Ashraf, S. S., Sochacka, E., & Cain, R. (1999). Single atom modification (O–>S) of tRNA confers ribosome binding. RNA, 5, 188–194.
Wei, F. Y., Suzuki, T., Watanabe, S., Kimura, S., Kaitsuka, T., Fujimura, A., Matsui, H., Atta, M., Michiue, H., Fontecave, M., Yamagata, K., & Suzuki, T. (2011). Tomizawa, K., Deficit of tRNALys modification by Cdkal1 causes the development of type 2 diabetes in mice. The. Journal of Clinical Investigation, 121(9), 3598–3608.
Yarian, C., Townsend, H., Czestkowski, W., Sochacka, E., Malkiewicz, A. J., Guenther, R., Miskiewicz, A., & Agris, P. F. (2002). Accurate translation of the genetic code depends on tRNA modified nucleosides. The Journal of Biological Chemistry, 277(19), 16391–16395.
Matuszewski, M., Wojciechowski, J., Miyauchi, K., Gdaniec, Z., Wolf, W. M., Suzuki, T., & Sochacka, E. A. (2017). Hydantoin isoform of cyclic N6-threonylcarbamoyladenosine (ct6A) is present in tRNAs. Nucleic Acids Research, 45(4), 2137–2149.
Hehre, J., Radom, W. L., Schleyer, P. V. R., & Pople, J. A. (1986). Ab initio molecular orbital theory (pp. 4. New York: Wiley).
Kumbhar, N. M., & Sonawane, K. D. (2011). Iso-energetic multiple conformations of hypermodified nucleic acid base wybutine (yW) which occur at 37th position in anticodon loop of tRNAPhe. Journal of Molecular Graphics and Modelling, 29, 935–946.
Sonavane, U. B., Sonawane, K. D., & Tewari, R. (2002). Conformational preferences of base substituent in hypermodified nucleotide Queuosine 5’-monophosphate ‘pQ’ and protonated variant ‘pQH+’. Journal of Biomolecular Structure and Dynamics, 20(3), 473–485.
Pullman, B., & Pullman, A. (1974). Molecular orbital calculations on the conformation of amino acid residues of proteins. Advances in Protein Chemistry, 28, 347–526.
Tewari, R. (1987). Theorotical studies on conformational preferences of modified nucleic acid base N6-(N-glycylcarbonyl) Adenine. International Journal of Quantum Chemistry, XXX1, 611–623.
Sonavane, U. B., Sonawane, K. D., Morin, A., Grosjean, H., & Tewari, R. (1999). N(7)- Protonation induced conformational flipping in hypermodified nucleic acid bases N6-(N-threonylcarbonyl) adenine and its 2-Methylthio- or N(6)-methyl-derivatives. International Journal of Quantum Chemistry, 75, 223–229.
Francl, M. M., Pietro, W. J., Hehre, W. J., Binkley, J. S., DeFrees, D. J., Pople, J. A., & Gordon, M. S. (1982). Self-Consistent Molecular Orbital Methods. 23. A polarization-type basis set for 2nd-row elements. The Journal of Chemical Physics, 77, 3654–65.
Frisch, M., Scalmani, G., Vreven, T., & Zheng, G. (2009). Analytic second derivatives for semiempirical models based on MNDO. Molecular Physics, 107, 881–887.
Cornell, W. D., Cieplak, P., Bayly, C. I., & Kollman, P. A. (1993). Application of RESP charges to calculate conformational energies, hydrogen bond energies and free energies for solvation. Journal of American Chemical Society, 115, 9620–9631.
Bayly, C. I., Ceiplak, P., Cornell, W. D., & Kollman, P. A. (1993). A well behaved electrostatic potential based method using charge restraints for deriving atomic charges: The RESP Model. Journal of Physical Chemistry, 97, 10269–10280.
Cieplak, P., Cornell, W. D., Bayly, C., & Kollman, P. A. (1995). Application of the Multimolecule and Multiconformational RESP Methodology to Biopolymers: Charge Derivation for DNA, RNA, and Proteins. Journal of Computational Chemistry, 16(11), 1357–1377.
Dupradeau, F. Y., Pigache, A., Zaffran, T., Savineau, C., Lelong, R., Grivel, N., Lelong, D., Rosanski, W., & Cieplak, P. (2010). The R.E.D. tools: advances in RESP and ESP charge deviation and force field library building. Physical Chemistry, Chemical Physics, 12, 7821–7839.
Ryckaert, J. P., Ciccotti, G., & Berendsen, H. J. C. (1977). Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. Journal of Computational Physics, 23, 327–341.
Berendsen, H. J. C., Postma, J. P. M., Gunsteren, W. F. V., & DiNola, A. H. J. R. (1984). Molecular dynamics with coupling to an external bath. Journal of Chemical Physics, 81(8), 3684–3690.
Debye, P. J. W. (1954). The collected papers of Peter J.W. Debye. Interscience publishers, Inc., New York.
Case, D. A., Cheatham, III, T. E., Darden, T., Gohlke, H., Luo, R., Merz, Jr, K. M., Onufriev, A., Simmerling, C., Wang, B., & Woods, R. J. (2005). The Amber Biomolecular Simulation Programs. Journal of Computational Chemistry, 26(16), 1668–1688.
Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., & Ferrin, T. E. (2004). UCSF Chimera -A visualization system for exploratory research and analysis. Journal of Computational Chemistry, 25(13), 1605–1612.
Sundaralingam, M. (1969). Stereochemistry of Nucleic Acids and Their Constituents.* IV. Allowed and Preferred Conformations of Nucleosides, Nucleoside Mono-, Di-, Tri-, Tetraphosphates. Nucleic Acids and Polynucleotide. Biopolymers, 7, 821–860.
Altona, C., & Sundaralingam, M. (1973). Conformational Analysis of the Sugar Ring in Nucleosides and Nucleotides. Improved Method for the Interpretation of Proton Magnetic Resonance Coupling Constants. Journal of the American Chemical Society, 95(7), 2333–2344.
Saenger, W. (1984). Defining Terms for the Nucleic Acids. In: Principles of Nucleic Acid Structure, Springer, New York, 9–28.
Cusack, S., Yaremchuk, A., & Tukalo, M. (1996). The crystal structures of T. Thermophilus lysyl-tRNA synthetase complexed with E. coli tRNALys and a T. thermophilus tRNALys transcript: anticodon recognition and conformational changes upon binding of a lysyl-adenylate analogue. The EMBO Journal, 15(22), 6321–6334.
Yokoyama, S., Watanabe, T., Murao, K., Ishikura, H., Yamaizumi, Z., Nishimura, S., & Miyazawa, T. (1985). Molecular mechanism of codon recognition by tRNA species with modified Uridine in the first position of the anticodon. Biochemistry, 82, 4905–1909.
Altona, C., & Sundaralingam, M. (1972). Conformational Analysis of the Sugar Ring in Nucleosides and Nucleotides. A New Description Using the Concept of Pseudorotation. Journal of American Chemical Society, 94(23), 8205–8212.
Clay, M. C., Ganser, L. R., Merriman, D. K., & Al-Hashimi, H. M. (2017). Resolving sugar puckers in excited state exposes slow modes of repuckering dynamics. Nucleic Acids Research, 45(14), e134.
Everaert, D. H., Peeters, O. M., De Ranter, C. J., Blaton, N. M., van Aerschot, A., & Herdewijin, P. (1993). Conformational analysis of substituent effects on the sugar puckering mode and the anti-HIV activity of 2’,3’-dideoxypyrimidine nucleosides. Antiviral Chemistry & Chemotherapy, 4(5), 289–299.
Aakeroy, C. B., Wijethunga, T. K., & Desper, J. (2015). Molecular electrostatic potential dependent selectivity of hydrogen bonding. New Journal of Chemistry, 39, 822.
Lescrinier, E., Nauwelaerts, K., Zanier, K., Poesen, K., Sattler, M., & Herdewijn, P. (2006). The naturally occurring N6-threonyl adenine in anticodon loop of Schizosaccharomyces pombe tRNAi causes formation of a unique U-turn motif. Nucleic Acids Research, 34(10), 2878–2886.
Acknowledgements
KDS gratefully acknowledges the financial support received from DST, New Delhi under the SERB-EMR project (EMR/2017/002688/BBM). ASD thankful to DST SERB for providing project fellowship. Authors are also thankful to Computer center, Shivaji University, Kolhapur for providing necessary facilities.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Dound, A.S., Fandilolu, P.M. & Sonawane, K.D. Structural Significance of Conformational Preferences and Ribose-Ring-Puckering of Hyper Modified Nucleotide 5’-Monophosphate 2-Methylthio Cyclic N6-Threonylcarbamoyladenosine (p-ms2ct6A) Present at 37th Position in Anticodon Loop of tRNALys. Cell Biochem Biophys 80, 665–680 (2022). https://doi.org/10.1007/s12013-022-01086-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12013-022-01086-0