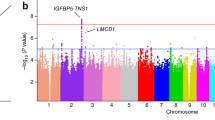
Abstract
Purpose of Review
Increases in the availability of genetic data and advances in the tools and methods for their analyses have enabled well-powered genetic association studies that have significantly enhanced our understanding of the genetic factors underlying both rare and common valve diseases. Valvular heart diseases, such as congenital valve malformations and degenerative valve lesions, increase the risk of heart failure, arrhythmias, and sudden death. In this review, we provide an updated overview of our current understanding of the genetic mechanisms underlying valvular heart diseases. With a focus on discoveries from the past 5 years, we describe recent insights into genetic risk and underlying biological pathways.
Recent Findings
Recently acquired knowledge around valvular heart disease genetics has provided important insights into novel mechanisms related to disease pathogenesis. Newly identified risk loci associated valvular heart disease mainly regulate the composition of the extracellular matrix, accelerate the endothelial-to-mesenchymal transition, contribute to cilia formation processes, and play roles in lipid metabolism.
Summary
Large-scale genomic analyses have identified numerous risk loci, genes, and biological pathways associated with degenerative valve disease and congenital valve malformations. Shared risk genes suggest common mechanistic pathways for various valve pathologies. More recent studies have combined cardiac magnetic resonance imaging and machine learning to offer a novel approach for exploring genotype-phenotype relationships regarding valve disease. Progress in the field holds promise for targeted prevention, particularly through the application of polygenic risk scores, and innovative therapies based on the biological mechanisms for predominant forms of valvular heart diseases.

Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Go AS, Mozaffarian D, Roger VL, Benjamin EJ, Berry JD, Borden WB, et al. Executive summary: heart disease and stroke statistics–2013 update: a report from the American Heart Association. Circulation. 2013;127(1):143–52. https://doi.org/10.1161/CIR.0B013E318282AB8F.
Nkomo VT, Gardin JM, Skelton TN, Gottdiener JS, Scott CG, Enriquez-Sarano M. Burden of valvular heart diseases: a population-based study. Lancet. 2006;368(9540):1005–1011. https://doi.org/10.1016/S0140-6736(06)69208-8.
Virani SS, Alonso A, Benjamin EJ, Bittencourt MS, Callaway CW, Carson AP, et al. Heart disease and stroke statistics-2020 update: a report from the American Heart Association. Circulation. 2020;141(9):e139-e596. https://doi.org/10.1161/CIR.0000000000000757.
Yang Y, Wang Z, Chen Z, Wang X, Zhang L, Li S, et al. Current status and etiology of valvular heart disease in China: a population-based survey. BMC Cardiovasc Disord. 2021;21(1):339. https://doi.org/10.1186/S12872-021-02154-8.
Cripe L, Andelfinger G, Martin LJ, Shooner K, Benson D. Bicuspid aortic valve is heritable. J Am Coll Cardiol. 2004;44(1):138–43. https://doi.org/10.1016/J.JACC.2004.03.050.
Kodali SK, Velagapudi P, Hahn RT, Abbott D, Leon MB. Valvular heart disease in patients ≥80 years of age. J Am Coll Cardiol. 2018;71(18):2058-2072. https://doi.org/10.1016/J.JACC.2018.03.459.
Delling FN, Rong J, Larson MG, Lehman B, Osypiuk E, Stantchev P, et al. Familial clustering of mitral valve prolapse in the community. Circulation. 2015;131(3):263-268. https://doi.org/10.1161/CIRCULATIONAHA.114.012594.
Probst V, Le Scouarnec S, Legendre A, Jousseaume V, Jaafar P, Nguyen JM, et al. Familial aggregation of calcific aortic valve stenosis in the western part of France. Circulation. 2006;113(6):856-860. https://doi.org/10.1161/CIRCULATIONAHA.105.569467.
Andell P, Li X, Martinsson A, Andersson C, Stagmo M, Zöller B, et al. Epidemiology of valvular heart disease in a Swedish nationwide hospital-based register study. Heart. 2017;103(21):1696-1703. https://doi.org/10.1136/HEARTJNL-2016-310894.
• Blaser MC, Kraler S, Lüscher TF, Aikawa E. Network-guided multiomic mapping of aortic valve calcification. Arterioscler Thromb Vasc Biol. 2023;43(3):417–26. https://doi.org/10.1161/ATVBAHA.122.318334. In this review, the authors cite recent efforts to apply epigenomics, transcriptomics, proteomic, and metabolic initiatives to study aortic valve calcification.
•• Moncla L-HM, Briend M, Bossé Y, Mathieu P. Calcific aortic valve disease: mechanisms, prevention and treatment. Nat Rev Cardiol. 2023;20(8):546–59. https://doi.org/10.1038/s41569-023-00845-7. This review provides a comprehensive summary of the risk factors, genetics, and molecular mechanisms involved in CAVD.
Martinsson A, Li X, Zöller B, Andell P, Andersson C, Sundquist K, et al. Familial aggregation of aortic valvular stenosis: a nationwide study of sibling risk. Circ Cardiovasc Genet. 2017;10(6):e001742. https://doi.org/10.1161/CIRCGENETICS.117.001742/-/DC1
Boureau AS, Karakachoff M, Le Scouarnec S, Capoulade R, Cueff C, de Decker L, et al. Heritability of aortic valve stenosis and bicuspid enrichment in families with aortic valve stenosis. Int J Cardiol. 2022;359:91–8. https://doi.org/10.1016/j.ijcard.2022.04.022.
Thanassoulis G, Campbell CY, Owens D, Smith J, Smith AV, Peloso G, et al. Genetic associations with valvular calcification and aortic stenosis. N Engl J Med. 2013;368(6):503–12. https://doi.org/10.1056/NEJMoa1109034.
Rogers MA, Atkins SK, Zheng KH, Singh SA, Chelvanambi S, Pham TH, et al. Lipoprotein(a) induces vesicular cardiovascular calcification revealed with single-extracellular vesicle analysis. Front Cardiovasc Med. 2022;9:778919. https://doi.org/10.3389/FCVM.2022.778919/BIBTEX.
Nazarzadeh M, Pinho-Gomes A-C, Bidel Z, Dehghan A, Canoy D, Hassaine A, et al. Plasma lipids and risk of aortic valve stenosis: a Mendelian randomization study. Eur Heart J. 2020;41:3913–20. https://doi.org/10.1093/eurheartj/ehaa070. This Mendelian randomization-based study provided robust support for the causal association link between plasma lipids and AVS.
Sanderson E, Glymour MM, Holmes MV, Kang H, Morrison J, Munafò MR, et al. Mendelian randomization. Nat Rev Methods Primers. 2022;2(1):6. https://doi.org/10.1038/s43586-021-00092-5.
Langsted A, Nordestgaard BG, Benn M, Tybjærg-Hansen A, Kamstrup PR. PCSK9 R46L loss-of-function mutation reduces lipoprotein(a), LDL cholesterol, and risk of aortic valve stenosis. J Clin Endocrinol Metab. 2016;101(9):3281–7. https://doi.org/10.1210/JC.2016-1206.
Perrot N, Valerio V, Moschetta D, Boekholdt SM, Dina C, Chen HY, et al. Genetic and in vitro inhibition of PCSK9 and calcific aortic valve stenosis. JACC Basic Transl Sci. 2020;5(7):649–61. https://doi.org/10.1016/j.jacbts.2020.05.004.
Helgadottir A, Thorleifsson G, Gretarsdottir S, Stefansson OA, Tragante V, Thorolfsdottir RB, et al. Genome-wide analysis yields new loci associating with aortic valve stenosis. Nat Commun. 2018;9(1):987. https://doi.org/10.1038/s41467-018-03252-6.
Thériault S, Gaudreault N, Lamontagne M, Rosa M, Boulanger MC, Messika-Zeitoun D, et al. A transcriptome-wide association study identifies PALMD as a susceptibility gene for calcific aortic valve stenosis. Nat Commun. 2018;9(1):988. https://doi.org/10.1038/s41467-018-03260-6.
•• Small AM, Peloso G, Linefsky J, Aragam J, Galloway A, Tanukonda V, et al. Multiancestry genome-wide association study of aortic stenosis identifies multiple novel loci in the million veteran program. Circulation. 2023;147(12):942–55. https://doi.org/10.1161/circulationaha.122.061451. This study is one of the largest and most ancestrally diverse GWAS of AS, which identified 14 risk loci located in 11 unique genomic regions including 5 previously reported loci and 6 new loci. Two of the loci have been replicated in Hispanic and Black individuals.
Wang S, Yu H, Gao J, Chen J, He P, Zhong H, et al. PALMD regulates aortic valve calcification via altered glycolysis and NF-κB-mediated inflammation. J Biol Chem. 2022;298(5):101887. https://doi.org/10.1016/j.jbc.2022.101887.
Sun JY, Hua Y, Shen H, Qu Q, Kan JY, Kong XQ, et al. Identification of key genes in calcific aortic valve disease via weighted gene co-expression network analysis. BMC Med Genomics. 2021;14(1):135. https://doi.org/10.1186/S12920-021-00989-W/FIGURES/6.
Chen HY, Cairns BJ, Small AM, Burr HA, Ambikkumar A, Martinsson A, et al. Association of FADS1/2 locus variants and polyunsaturated fatty acids with aortic stenosis. JAMA Cardiol. 2020;5(6):694–702. https://doi.org/10.1001/JAMACARDIO.2020.0246. This GWAS provided the first evidence for the genetic role of FADS1/2 involved in ω-6 and ω-3 fatty acid biosynthesis in the risk of AS and calcification.
Surendran A, Edel A, Chandran M, Bogaert P, Hassan-Tash P, Asokan AK, et al. Metabolomic signature of human aortic valve stenosis. JACC Basic to translational science. 2020;5(12):1163–77. https://doi.org/10.1016/j.jacbts.2020.10.001.
Massera D, Kizer JR, Dweck MR. Mechanisms of mitral annular calcification. Trends Cardiovasc Med. 2020;30(5):289–95. https://doi.org/10.1016/J.TCM.2019.07.011.
Patlolla SH, Schaff HV, Nishimura RA, Geske JB, Lahr BD, Lee AT, et al. Mitral annular calcification in obstructive hypertrophic cardiomyopathy: prevalence and outcomes. Ann Thorac Surg. 2022;114(5):1679–87. https://doi.org/10.1016/j.athoracsur.2021.09.077.
Kataria R, Castagna F, Madan S, Kim P, Saeed O, Adjepong YA, et al. Severity of functional mitral regurgitation on admission for acute decompensated heart failure predicts long-term risk of rehospitalization and death. J Am Heart Assoc. 2022;11(1):e022908. https://doi.org/10.1161/JAHA.121.022908.
Flint N, Raschpichler M, Rader F, Shmueli H, Siegel RJ. Asymptomatic degenerative mitral regurgitation: a review. JAMA Cardiol. 2020;5(3):346-355. https://doi.org/10.1001/JAMACARDIO.2019.5466.
Kyndt F, Gueffet JP, Probst V, Jaafar P, Legendre A, Le Bouffant F, et al. Mutations in the gene encoding filamin A as a cause for familial cardiac valvular dystrophy. Circulation. 2007;115(1):40–49. https://doi.org/10.1161/CIRCULATIONAHA.106.622621.
Kyndt F, Schott JJ, Trochu JN, Baranger F, Herbert O, Scott V, et al. Mapping of X-linked myxomatous valvular dystrophy to chromosome Xq28. Am J Hum Genet. 1998;62(3):627–632. https://doi.org/10.1086/301747.
Deng W, Lopez-Camacho C, Tang JY, Mendoza-Villanueva D, Maya-Mendoza A, Jackson DA, et al. Cytoskeletal protein filamin A is a nucleolar protein that suppresses ribosomal RNA gene transcription. Proc Natl Acad Sci U S A. 2012;109(5):1524-1529. https://doi.org/10.1073/PNAS.1107879109/-/DCSUPPLEMENTAL/PNAS.201107879SI.PDF.
Le Tourneau T, Le Scouarnec S, Cueff C, Bernstein D, Aalberts JJJ, Lecointe S, et al. New insights into mitral valve dystrophy: a Filamin-A genotype–phenotype and outcome study. Eur Heart J. 2018;39(15):1269-1277. https://doi.org/10.1093/EURHEARTJ/EHX505.
Toomer KA, Yu M, Fulmer D, Moore KS, Moore R, et al. Primary cilia defects causing mitral valve prolapse. Sci Transl Med. 2019;11(493):eaax0290. https://doi.org/10.1126/SCITRANSLMED.AAX0290.
Dina C, Bouatia-Naji N, Tucker N, Delling FN, Toomer K, Durst R, et al. Genetic association analyses highlight biological pathways underlying mitral valve prolapsed. Nat Genet. 2015;47(10):1206-1211. https://doi.org/10.1038/ng.3383.
• Kyryachenko S, Georges A, Yu M, Barrandou T, Guo L, Bruneval P, et al. Chromatin accessibility of human mitral valves and functional assessment of MVP risk loci. Circ Res. 2021;128(5):e84–101. https://doi.org/10.1161/CIRCRESAHA.120.317581. This article described unprecedented genome-wide open chromatin profiles from human pathogenic and nonpathogenic MVs.
Yu M, Georges A, Tucker NR, Kyryachenko S, Toomer K, Schott JJ, et al. Genome-wide association study-driven gene-set analyses, genetic, and functional follow-up suggest Glis1 as a susceptibility gene for mitral valve prolapse. Circ Genom Precis Med. 2019;12(5):e002497. https://doi.org/10.1161/CIRCGEN.119.002497
•• Roselli C, Yu M, Nauffal V, Georges A, Yang Q, Love K, et al. Genome-wide association study reveals novel genetic loci: a new polygenic risk score for mitral valve prolapse. Eur Heart J. 2022;43(17):1668–80. https://doi.org/10.1093/eurheartj/ehac049. This study is the largest GWAS published for MVP to date, which identified 14 risk loci, including those previously found and new genes, as well as presented the first application of PRS assessment in MVP.
Cahill TJ, Prothero A, Wilson J, Kennedy A, Brubert J, Masters M, et al. Community prevalence, mechanisms and outcome of mitral or tricuspid regurgitation. Heart. 2021;107(12):1003-1009. https://doi.org/10.1136/HEARTJNL-2020-318482.
Hahn RT, Weckbach LT, Noack T, Hamid N, Kitamura M, Bae R, et al. Proposal for a standard echocardiographic tricuspid valve nomenclature. JACC Cardiovasc Imaging. 2021;14(7):1299–1305. https://doi.org/10.1016/J.JCMG.2021.01.012.
Tian C, Yang Y, Ke Y, Yang L, Zhong L, Wang Z, et al. Integrative analyses of genes associated with right ventricular cardiomyopathy induced by tricuspid regurgitation. Front Genet. 2021;12:708275. https://doi.org/10.3389/FGENE.2021.708275/BIBTEX.
Stephensen SS, Sigfusson G, Eiriksson H, Sverrisson JT, Torfason B, Haraldsson A, et al. Congenital cardiac malformations in Iceland from 1990 through 1999. Cardiol Young. 2004;14(4):396–401. https://doi.org/10.1017/S1047951104004081.
Galian-Gay L, Carro Hevia A, Teixido-Tura G, Rodriguez Palomares J, Gutierrez-Moreno L, Maldonado G, et al. Familial clustering of bicuspid aortic valve and its relationship with aortic dilation in first-degree relatives. Heart. 2019;105(8):603-608. https://doi.org/10.1136/heartjnl-2018-313802.
• Debiec R, Hamby S, Jones P, Safwan KA, Sosin M, Hetherington S, et al. Contribution of NOTCH1 genetic variants to bicuspid aortic valve and other congenital lesions. Heart. 2022;108(14):1114–20. https://doi.org/10.1136/heartjnl-2021-320428. This study assessed NOTCH1 mutations in a large number of pedigrees and patient cohorts and estimated these genes to be causal in 2% of familial and <0.1% of sporadic non-syndromic BAV.
Debiec R, Hamby SE, Jones PD, Coolman S, Asiani M, Kharodia S, et al. Novel loss of function mutation in NOTCH1 in a family with bicuspid aortic valve, ventricular septal defect, thoracic aortic aneurysm, and aortic valve stenosis. Mol Genet Genomic Med. 2020;8(10):e1437. https://doi.org/10.1002/MGG3.1437
Xu YJ, Di RM, Qiao Q, Li XM, Huang RT, Xue S, et al. GATA6 loss-of-function mutation contributes to congenital bicuspid aortic valve. Gene. 2018;663:115–120. https://doi.org/10.1016/J.GENE.2018.04.018.
Li RG, Xu YJ, Wang J, Liu XY, Yuan F, Huang RT, et al. GATA4 loss-of-function mutation and the congenitally bicuspid aortic valve. Am J Cardiol. 2018;121(4):469–474. https://doi.org/10.1016/j.amjcard.2017.11.012.
Gharibeh L, Komati H, Bossé Y, Boodhwani M, Heydarpour M, Fortier M, et al. GATA6 regulates aortic valve remodeling, and its haploinsuffciency leads to right-left type bicuspid aortic valve. Circulation. 2018;138(10):1025–1038. https://doi.org/10.1161/CIRCULATIONAHA.117.029506.
Gould RA, Aziz H, Woods CE, Seman-Senderos MA, Sparks E, Preuss C, et al. ROBO4 variants predispose individuals to bicuspid aortic valve and thoracic aortic aneurysm. Nat Genet. 2018;51(1):42–50. https://doi.org/10.1038/s41588-018-0265-y.
Jaouadi H, Gérard H, Théron A, Collod-Béroud G, Collart F, Avierinos JF, et al. Identification of non-synonymous variations in ROBO1 and GATA5 genes in a family with bicuspid aortic valve disease. J Hum Genet. 2022;67(9):515–518. https://doi.org/10.1038/s10038-022-01036-x.
Theis JL, Niaz T, Sundsbak RS, Fogarty ZC, Bamlet WR, Hagler DJ, et al. CELSR1 risk alleles in familial bicuspid aortic valve and hypoplastic left heart syndrome. Circ Genom Precis Med. 2022;15(2):e003523. https://doi.org/10.1161/CIRCGEN.121.003523.
Fulmer D, Toomer K, Guo L, Moore K, Glover J, Moore R, et al. Defects in the exocyst-cilia machinery cause bicuspid aortic valve disease and aortic stenosis. Circulation. 2019;140(16):1331–1341. https://doi.org/10.1161/CIRCULATIONAHA.119.038376.
Gehlen J, Stundl A, Debiec R, Fontana F, Krane M, Sharipova D, et al. Elucidation of the genetic causes of bicuspid aortic valve disease. Cardiovasc Res. 2023;119(3):857–866. https://doi.org/10.1093/CVR/CVAC099.
Yang B, Zhou W, Jiao J, Nielsen JB, Mathis MR, Heydarpour M, et al. Protein-altering and regulatory genetic variants near GATA4 implicated in bicuspid aortic valve. Nat Commun. 2017;8:15481. https://doi.org/10.1038/ncomms15481.
Xiong T-Y, Liu C, Liao Y-B, Zheng W, Li Y-J, Li X, et al. Differences in metabolic profiles between bicuspid and tricuspid aortic stenosis in the setting of transcatheter aortic valve replacement. BMC Cardiovasc Disord. 2020;20(1):229. https://doi.org/10.1186/s12872-020-01491-4.
O’Donnell A, Yutz KE. Mechanisms of heart valve development and disease. Development (Cambridge). 2020;147. https://doi.org/10.1242/DEV.183020/224244.
Wang C, Li Y, Lv J, Jin J, Hu X, Kuang X, et al. Recommendation for cardiac magnetic resonance imaging-based phenotypic study: Imaging part. Phenomics. 2021;1(14):151–170. https://doi.org/10.1007/S43657-021-00018-X.
Córdova-Palomera A, Tcheandjieu C, Fries JA, Varma P, Chen VS, Fiterau M, et al. Cardiac imaging of aortic valve area from 34 287 UK Biobank participants reveals novel genetic associations and shared genetic comorbidity with multiple disease phenotypes. Circ Genom Precis Med. 2020;13(6):e003014. https://doi.org/10.1161/CIRCGEN.120.003014.
• Yu M, Tcheandjieu C, Georges A, Xiao K, Tejeda H, Dina C, et al. Computational estimates of annular diameter reveal genetic determinants of mitral valve function and disease. JCI Insight. 2022;7(3):e146580. https://doi.org/10.1172/jci.insight.146580Based on the automated estimates of mitral valve annular diameter from MRI images from the UK Biobank, this study identified 10 risk loci, including GOSR2. The polygenic scoring of MV annular diameter in systole was predictive of risk MVP.
Funding
This work was sponsored by the Shanghai Sailing Program (21YF1452900) and Shanghai Clinical Research Program (20234Y0239) to Dr Yu. NBN is supported by the European Research Council grant (ERC-Stg-ROSALIND-716628), the French Society of Cardiology through Fondation “Coeur et Recherche,” “La Fédération Française de Cardiologie”, and Fondation pour la Recherche Medicale (FRM-2023).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors confirm that there is no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yu, M., Bouatia-Naji, N. Insights into the Inherited Basis of Valvular Heart Disease. Curr Cardiol Rep (2024). https://doi.org/10.1007/s11886-024-02041-6
Accepted:
Published:
DOI: https://doi.org/10.1007/s11886-024-02041-6