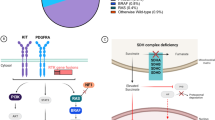
Opinion statement
Gastrointestinal stromal tumor (GIST) constitutes a paradigm for clinically effective targeted inhibition of oncogenic driver mutations. Therefore, GIST has emerged as a compelling clinical and biological model to study oncogene addiction and to validate preclinical concepts for drug response and drug resistance. Oncogenic activation of KIT or PDGFRA receptor tyrosine kinases is the essential drivers of GIST progression throughout all stages of the disease. Interestingly, KIT/PDGFRA genotype predicts the response to first-line imatinib and to all tyrosine kinase inhibitors (TKIs) approved or in investigation after imatinib failure. Considering that TKIs are effective only against a subset of KIT or PDGFRA resistance mutations, close monitoring of tumor dynamics with non-invasive methods such as liquid biopsy emerges as a necessary step forward in the field. Liquid biopsy, in contrast to solid tumor biopsy, aims to characterize tumors irrespective of heterogeneity. Although there are several components in the peripheral blood, most recent studies have been focused on circulating tumor (ct)DNA, due to the technological feasibility, the stability of DNA itself and DNA alterations, and the therapeutic development in precision oncology largely based on the identification of genetic driver mutations. In the present review, we systematically dissect the current wealth of data of ctDNA in GIST. To do so, a critical understanding of the promises and limitations of the current technologies will be followed by an exposition of the knowledge gathered with such studies in GIST. Collectively, our goal is to establish clear premises that can be used as the foundations to build future studies towards the clinical implementation of ctDNA evaluation in GIST patients.

Similar content being viewed by others
References and Recommended Reading
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Dienstmann R, Tabernero J. A precision approach to tumour treatment. Nature. 2017;548:40–1.
Schwartzberg L, Kim ES, Liu D, Schrag D. Precision oncology: who, how, what, when, and when not? Am Soc Clin Oncol Educ Book. 2017;37:160–9.
Chakravarty D, Gao J, Phillips SM, et al. OncoKB: a precision oncology knowledge base. JCO Precis Oncol. 2017. https://doi.org/10.1200/PO.17.00011.
Morash M, Mitchell H, Beltran H, Elemento O, Pathak J. The role of next-generation sequencing in precision medicine: a review of outcomes in oncology. J Pers Med. 2018. https://doi.org/10.3390/jpm8030030.
Sicklick JK, Kato S, Okamura R, et al. Molecular profiling of cancer patients enables personalized combination therapy: the I-PREDICT study. Nat Med. 2019;25:744–50.
Tamborero D, Dienstmann R, Rachid MH, et al. Support systems to guide clinical decision-making in precision oncology: The Cancer Core Europe Molecular Tumor Board Portal. Nat Med. 2020;26:992–94.
FGM 2025 Workflow Study Group (Alliance nationale des Sciences de la Vie et de la Santé), Auzanneau C, Bacq D, et al. Feasibility of high-throughput sequencing in clinical routine cancer care: lessons from the cancer pilot project of the France Genomic Medicine 2025 plan. ESMO Open. 2020. https://doi.org/10.1136/esmoopen-2020-000744.
Karlovich CA, Williams PM. Clinical applications of next-generation sequencing in precision oncology. Cancer J Sudbury Mass. 2019;25:264–71.
Kato S, Kim KH, Lim HJ, et al. Real-world data from a molecular tumor board demonstrates improved outcomes with a precision N-of-One strategy. Nat Commun. 2020;11:4965.
Middleton G, Fletcher P, Popat S, et al. The National Lung Matrix Trial of personalized therapy in lung cancer. Nature. 2020;583:807–12.
The cost of sequencing a human genome. In: Genome.gov. https://www.genome.gov/about-genomics/fact-sheets/Sequencing-Human-Genome-cost. Accessed 25 Sep 2020
Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24.
Li MM, Datto M, Duncavage EJ, et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer. J Mol Diagn JMD. 2017;19:4–23.
Jennings LJ, Arcila ME, Corless C, Kamel-Reid S, Lubin IM, Pfeifer J, et al. Guidelines for validation of next-generation sequencing–based oncology panels. J Mol Diagn. 2017;19:341–65.
Heitzer E, Haque IS, Roberts CES, Speicher MR. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat Rev Genet. 2019;20:71–88.
Ravegnini G, Sammarini G, Serrano C, Nannini M, Pantaleo MA, Hrelia P, et al. Clinical relevance of circulating molecules in cancer: focus on gastrointestinal stromal tumors. Ther Adv Med Oncol. 2019;11:175883591983190.
Demetri GD, von Mehren M, Antonescu CR, et al. NCCN Task Force report: update on the management of patients with gastrointestinal stromal tumors. J Natl Compr Cancer Netw JNCCN. 2010;8(Suppl 2):S1–41 quiz S42–4.
Ducimetiére F, Lurkin A, Ranchére-Vince D, et al. Incidence of sarcoma histotypes and molecular subtypes in a prospective epidemiological study with central pathology review and molecular testing. PLoS One. 2011;6:e20294.
Fletcher CDM, Berman JJ, Corless C, et al. Diagnosis of gastrointestinal stromal tumors: a consensus approach. Hum Pathol. 2002;33:459–65.
Goettsch WG, Bos SD, Breekveldt-Postma N, Casparie M, Herings RMC, Hogendoorn PCW. Incidence of gastrointestinal stromal tumours is underestimated: results of a nation-wide study. Eur J Cancer. 2005;41:2868–72.
Hirota S, Isozaki K, Moriyama Y, et al. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science. 1998;279:577–80.
Corless CL, Barnett CM, Heinrich MC. Gastrointestinal stromal tumours: origin and molecular oncology. Nat Rev Cancer. 2011;11:865–78.
Corless CL, McGreevey L, Town A, Schroeder A, Bainbridge T, Harrell P, et al. KIT gene deletions at the intron 10-exon 11 boundary in GI stromal tumors. J Mol Diagn. 2004;6:366–70.
Yuzawa S, Opatowsky Y, Zhang Z, Mandiyan V, Lax I, Schlessinger J. Structural basis for activation of the receptor tyrosine kinase KIT by stem cell factor. Cell. 2007;130:323–34.
Lasota J, Corless CL, Heinrich MC, et al. Clinicopathologic profile of gastrointestinal stromal tumors (GISTs) with primary KIT exon 13 or exon 17 mutations: a multicenter study on 54 cases. Mod Pathol. 2008;21:476–84.
Hirota S, Ohashi A, Nishida T, Isozaki K, Kinoshita K, Shinomura Y, et al. Gain-of-function mutations of platelet-derived growth factor receptor α gene in gastrointestinal stromal tumors. Gastroenterology. 2003;125:660–7.
Heinrich MC, Corless CL, Duensing A, et al. PDGFRA activating mutations in gastrointestinal stromal tumors. Science. 2003;299:708–10.
Serrano C, George S, Valverde C, Olivares D, García-Valverde A, Suárez C, et al. Novel insights into the treatment of imatinib-resistant gastrointestinal stromal tumors. Target Oncol. 2017;12:277–88.
Demetri GD, von Mehren M, Blanke CD, et al. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N Engl J Med. 2002;347:472–80.
Heinrich MC, Corless CL, Demetri GD, et al. Kinase mutations and imatinib response in patients with metastatic gastrointestinal stromal tumor. J Clin Oncol. 2003;21:4342–9.
Liegl B, Kepten I, Le C, Zhu M, Demetri G, Heinrich M, et al. Heterogeneity of kinase inhibitor resistance mechanisms in GIST. J Pathol. 2008;216:64–74. This is the main study highlighting the presence of heterogeneity of secondary resistance mutations in GIST.
Serrano C, Fletcher JA. Overcoming heterogeneity in imatinib-resistant gastrointestinal stromal tumor. Oncotarget. 2019;10:6286–7.
Demetri GD, van Oosterom AT, Garrett CR, et al. Efficacy and safety of sunitinib in patients with advanced gastrointestinal stromal tumour after failure of imatinib: a randomised controlled trial. Lancet. 2006;368:1329–38.
Demetri GD, Reichardt P, Kang Y-K, et al. Efficacy and safety of regorafenib for advanced gastrointestinal stromal tumours after failure of imatinib and sunitinib (GRID): an international, multicentre, randomised, placebo-controlled, phase 3 trial. Lancet. 2013;381:295–302.
Nemunaitis J, Bauer S, Blay J-Y, et al. Intrigue: phase III study of ripretinib versus sunitinib in advanced gastrointestinal stromal tumor after imatinib. Future Oncol. 2019;16:4251–64.
Smith BD, Kaufman MD, Lu WP, et al. Ripretinib (DCC-2618) is a switch control kinase inhibitor of a broad spectrum of oncogenic and drug-resistant KIT and PDGFRA variants. Cancer Cell. 2019;35:738–51 e9.
Blay J-Y, Serrano C, Heinrich MC, et al. Ripretinib in patients with advanced gastrointestinal stromal tumours (INVICTUS): a double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol. 2020;21:923–34.
Grunewald S, Klug LR, Muhlenberg T, et al. Resistance to avapritinib in PDGFRA-driven GIST is caused by secondary mutations in the PDGFRA kinase domain. Cancer Discov. 2020. https://doi.org/10.1158/2159-8290.CD-20-0487.
Corless CL, Schroeder A, Griffith D, Town A, McGreevey L, Harrell P, et al. PDGFRA mutations in gastrointestinal stromal tumors: frequency, spectrum and in vitro sensitivity to imatinib. J Clin Oncol. 2005;23:5357–64.
Heinrich MC, Corless CL, Blanke CD, et al. Molecular correlates of imatinib resistance in gastrointestinal stromal tumors. J Clin Oncol. 2006;24:4764–74.
Heinrich MC, Jones RL, von Mehren M, et al. Avapritinib in advanced PDGFRA D842V-mutant gastrointestinal stromal tumour (NAVIGATOR): a multicentre, open-label, phase 1 trial. Lancet Oncol. 2020;21:935–46.
Serrano C, Wang Y, Mariño-Enríquez A, et al. KRAS and KIT gatekeeper mutations confer polyclonal primary imatinib resistance in GI stromal tumors: relevance of concomitant phosphatidylinositol 3-kinase/AKT dysregulation. J Clin Oncol. 2015;33:e93–6.
Said R, Guibert N, Oxnard GR, Tsimberidou AM. Circulating tumor DNA analysis in the era of precision oncology. Oncotarget. 2020;11:188–211.
Namløs HM, Boye K, Meza-Zepeda LA. Cell-free DNA in blood as a noninvasive insight into the sarcoma genome. Mol Asp Med. 2020;72:100827.
Li Q, Zhi X, Zhou J, Tao R, Zhang J, Chen P, et al. Circulating tumor cells as a prognostic and predictive marker in gastrointestinal stromal tumors: a prospective study. Oncotarget. 2016;7:36645–54.
Atay S, Banskota S, Crow J, Sethi G, Rink L, Godwin AK. Oncogenic KIT-containing exosomes increase gastrointestinal stromal tumor cell invasion. Proc Natl Acad Sci. 2014;111:711–6.
Junquera C, Castiella T, Muñoz G, Fernández-Pacheco R, Luesma MJ, Monzón M. Biogenesis of a new type of extracellular vesicles in gastrointestinal stromal tumors: ultrastructural profiles of spheresomes. Histochem Cell Biol. 2016;146:557–67.
Atay S, Wilkey DW, Milhem M, Merchant M, Godwin AK. Insights into the Proteome of Gastrointestinal Stromal Tumors-Derived Exosomes Reveals New Potential Diagnostic Biomarkers. Mol Cell Proteomics. 2018;17:495–515.
Demetri GD, Jeffers M, Reichardt P, et al. Mutational analysis of plasma DNA from patients (pts) in the phase III GRID study of regorafenib (REG) versus placebo (PL) in tyrosine kinase inhibitor (TKI)-refractory GIST: correlating genotype with clinical outcomes. J Clin Oncol. 2013;31:10503.
Maier J, Lange T, Kerle I, et al. Detection of mutant free circulating tumor DNA in the plasma of patients with gastrointestinal stromal tumor harboring activating mutations of CKIT or PDGFRA. Clin Cancer Res. 2013;19:4854–67.
Yoo C, Ryu M-H, Na YS, Ryoo B-Y, Park SR, Kang Y-K. Analysis of serum protein biomarkers, circulating tumor DNA, and dovitinib activity in patients with tyrosine kinase inhibitor-refractory gastrointestinal stromal tumors. Ann Oncol Off J Eur Soc. Med Oncol. 2014;25:2272–7.
Bauer S, Herold T, Mühlenberg T, Reis A-C, Falkenhorst J, Backs M, et al. Plasma sequencing to detect a multitude of secondary KIT resistance mutations in metastatic gastrointestinal stromal tumors (GIST). J Clin Oncol. 2015;33:–10518.
Kang G, Bae BN, Sohn BS, Pyo J-S, Kang GH, Kim K-M. Detection of KIT and PDGFRA mutations in the plasma of patients with gastrointestinal stromal tumor. Target Oncol. 2015;10:597–601.
Kang G, Sohn BS, Pyo J-S, Kim JY, Lee B, Kim K-M. Detecting primary KIT mutations in presurgical plasma of patients with gastrointestinal stromal tumor. Mol Diagn Ther. 2016;20:347–51.
Wada N, Kurokawa Y, Takahashi T, et al. Detecting secondary C-KIT mutations in the peripheral blood of patients with imatinib-resistant gastrointestinal stromal tumor. Oncology. 2016;90:112–7.
Boonstra PA, ter Elst A, Tibbesma M, et al. A single digital droplet PCR assay to detect multiple KIT exon 11 mutations in tumor and plasma from patients with gastrointestinal stromal tumors. Oncotarget. 2018;9:13870–83.
Namløs HM, Boye K, Mishkin SJ, et al. Noninvasive detection of ctDNA reveals intratumor heterogeneity and is associated with tumor burden in gastrointestinal stromal tumor. Mol Cancer Ther. 2018;17:2473–80.
Xu H, Chen L, Shao Y, Zhu D, Zhi X, Zhang Q, et al. Clinical application of circulating tumor DNA in the genetic analysis of patients with advanced GIST. Mol Cancer Ther. 2018;17:290–6.
• Jilg S, Rassner M, Maier J, et al. Circulating cKIT and PDGFRA DNA indicates disease activity in Gastrointestinal Stromal Tumor (GIST). Int J Cancer. 2019;145:2292–303 Study on ctDNA in GIST using two different technologies: ddPCR and AS-PCR.
Serrano C, Leal A, Kuang Y, et al. Phase I study of rapid alternation of sunitinib and regorafenib for the treatment of tyrosine kinase inhibitor refractory gastrointestinal stromal tumors. Clin Cancer Res. 2019;25:7287–93 Study on ctDNA in GIST using two different technologies: ddPCR and TEC-seq.
Arshad J, Roberts A, Ahmed J, Cotta J, Pico BA, Kwon D, et al. Utility of circulating tumor DNA in the management of patients with GI stromal tumor: analysis of 243 patients. JCO Precis Oncol. 2020:66–73. Largest series of GIST patients evaluated by ctDNA in GIST. First report of successful therapeutic intervention following ctDNA assessment.
• Serrano C, Vivancos A, López-Pousa A, et al. Clinical value of next generation sequencing of plasma cell-free DNA in gastrointestinal stromal tumors. BMC Cancer. 2020;20:99 Study on ctDNA in GIST using two different technologies: ddPCR and amplicon-seq.
Merker JD, Oxnard GR, Compton C, et al. Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J Clin Oncol. 2018;36:1631–41.
Gerber T, Taschner-Mandl S, Saloberger-Sindhöringer L, et al. Assessment of pre-analytical sample handling conditions for comprehensive liquid biopsy analysis. J Mol Diagn. 2020;22:1070–86.
Johansson G, Andersson D, Filges S, Li J, Muth A, Godfrey TE, et al. Considerations and quality controls when analyzing cell-free tumor DNA. Biomol Detect Quantif. 2019;17:100078.
Markus H, Contente-Cuomo T, Farooq M, et al. Evaluation of pre-analytical factors affecting plasma DNA analysis. Sci Rep. 2018;8:7375.
Nikolaev S, Lemmens L, Koessler T, Blouin J-L, Nouspikel T. Circulating tumoral DNA: preanalytical validation and quality control in a diagnostic laboratory. Anal Biochem. 2018;542:34–9.
Godsey JH, Consortium on behalf of the B, Silvestro A, et al. Generic protocols for the analytical validation of next-generation sequencing-based ctDNA assays: A Joint Consensus Recommendation of the BloodPAC’s Analytical Variables Working Group. Clin Chem. 2020;66:1156–66.
Torga G, Pienta KJ. Patient-paired sample congruence between 2 commercial liquid biopsy tests. JAMA Oncol. 2018;4:868–70.
Torga G, Pienta KJ. Regarding the congruence between 2 circulating tumor DNA sequencing assays—reply. JAMA Oncol. 2018;4:1431.
Nong J, Gong Y, Guan Y, et al. Circulating tumor DNA analysis depicts subclonal architecture and genomic evolution of small cell lung cancer. Nat Commun. 2018;9:3114.
Murtaza M, Dawson S-J, Pogrebniak K, et al. Multifocal clonal evolution characterized using circulating tumour DNA in a case of metastatic breast cancer. Nat Commun. 2015;6:8760.
•• Serrano C, Mariño-Enríquez A, Tao DL, et al. Complementary activity of tyrosine kinase inhibitors against secondary kit mutations in imatinib-resistant gastrointestinal stromal tumours. Br J Cancer. 2019;120:612–20 This is the first study showing in a comprehensive manner how all TKIs in GIST have activity against only a subset of KIT secondary mutations.
Serrano C, George S. Gastrointestinal stromal tumor: challenges and opportunities for a new decade. Clin Cancer Res. 2020;26:5078–85.
Newman AM, Lovejoy AF, Klass DM, et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol. 2016;34:547–55.
Gale D, Lawson ARJ, Howarth K, et al. Development of a highly sensitive liquid biopsy platform to detect clinically-relevant cancer mutations at low allele fractions in cell-free DNA. PLoS One. 2018;13:e0194630.
Ren Y, Zhang Y, Wang D, Liu F, Fu Y, Xiang S, et al. SinoDuplex: an improved duplex sequencing approach to detect low-frequency variants in plasma cfDNA samples. Genomics Proteomics Bioinformatics. 2020;18:81–90.
Razavi P, Li BT, Brown DN, et al. High-intensity sequencing reveals the sources of plasma circulating cell-free DNA variants. Nat Med. 2019;25:1928–37.
Funding
C.S. has received research funding from Karyopharm, Pfizer, Inc, Deciphera Pharmaceuticals, and Bayer AG; consulting fees (advisory role) from Immunicum AB, Deciphera Pharmaceuticals and Blueprint Medicines; payment for lectures from Bayer AG and Blueprint Medicines; and travel grants from Pharmamar, Pfizer, Bayer AG, Novartis and Lilly. D.P.J has received travel grants from Pfizer, Roemmers and Roche. The remaining authors declare no conflicts of interests
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
David Gómez-Peregrina declares that he has no conflict of interest. Alfonso García-Valverde declares that he has no conflict of interest. Daniel Pilco-Janeta has received travel grants from Pfizer, Roemmers, and Roche. César Serrano has received research funding (paid to his institution) from Karyopharm Therapeutics, Pfizer, Deciphera Pharmaceuticals, and Bayer AG; has received consulting fees (advisory role) from Immunicum AB, Deciphera Pharmaceuticals, and Blueprint Medicines; has received payment for lectures from Bayer AG and Blueprint Medicines; and has received travel grants from PharmaMar, Pfizer, Bayer AG, Novartis, and Lilly
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Sarcoma
Rights and permissions
About this article
Cite this article
Gómez-Peregrina, D., García-Valverde, A., Pilco-Janeta, D. et al. Liquid Biopsy in Gastrointestinal Stromal Tumors: Ready for Prime Time?. Curr. Treat. Options in Oncol. 22, 32 (2021). https://doi.org/10.1007/s11864-021-00832-5
Accepted:
Published:
DOI: https://doi.org/10.1007/s11864-021-00832-5